Team:Vilnius-Lithuania/Modeling
Modeling
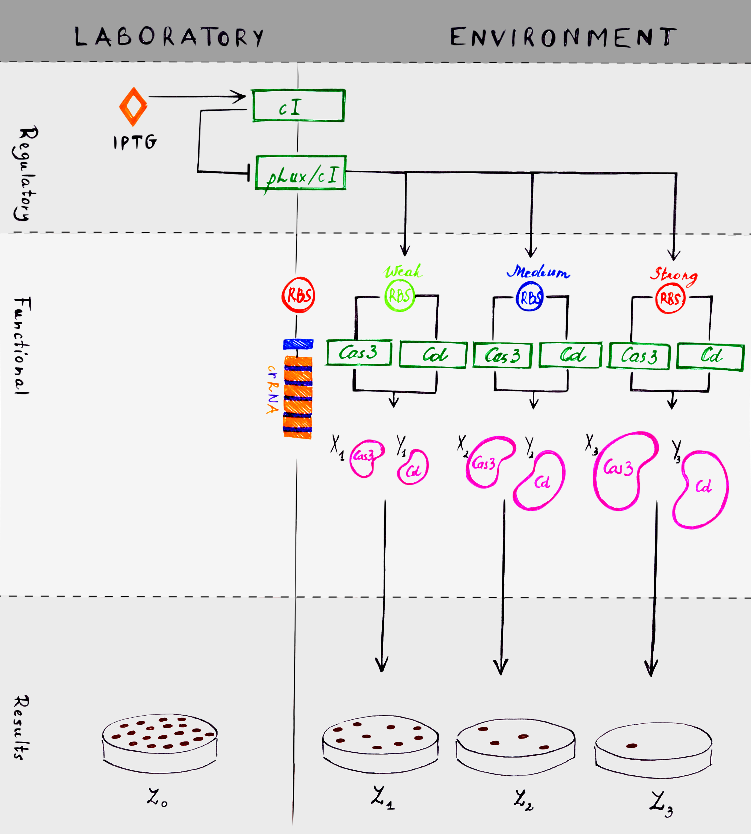
We created a scheme, which explains how our experiment should work. There are two moduls: laboratory and environment. System itself has two separate units, which interacts with each other. First of all, regulatory unit is resposible for system regulation and inductor IPTG is the main difference between laboratory and environment moduls. IPTG is not found in an environment, so that is why it is the main regulator. The second unit is functional, which does all the work to kill bacteria.
There are a few essential structures in the model, which are general for both moduls – laboratory and environment. These are cI repressor, pLux/cI promoter and crRNA. In the environmet cI expression is reduced, because there is no IPTG here. pLux/cI expression, on the other hand, is induced.
How everything works in a laboratory? When IPTG is added to the bacterial culture, it induces cI. cI repressor then repress pLux/cI promoter. Polymerase can not attach to the promoter and system is blocked. How everything would work in the environment? As we mentioned before, there is no IPTG here. So cI repressor production is not activated. Lack of cI means that pLux/cI promoter works and can activate Cas3 and Cascade transcription. Also positive loop occurs. LuxR and LuxI proteins interact with each other an forms a complex. This complex promotes pLux/cI promoter activation. We created three different options, because we added three different RBS sites – weak, medium and strong. Concentration of Cas3 and Cascade (Cd in the model) depends on these different RBS sites. If RBS is weak, smaller concentrations of Cas3 and Cd (X1 and Y1 respectively in the model) occurs. With medium RBS, Cas3 – X2 and Cd – Y2. With strong RBS, Cas3 – X3 and Cd – Y3. Relation between Cas3 concentrations (X1, X2 and X3) can be defined as an inequality:
How everything would work in the environment? As we mentioned before, there is no IPTG here. So cI repressor production is not activated. Lack of cI means that pLux/cI promoter works and can activate Cas3 and Cascade transcription. We created three different options, because we added three different RBS sites – weak, medium and strong. Concentration of Cas3 and Cascade (Cd in the model) depends on these different RBS sites.
X1 < X2 < X3
The same can be done with Cd concentrations:
Y1 < Y2 < Y3
The most interesting part, of course, are results. When bacteria grow in a lab and IPTG is given, they live and divide normaly. In the environment there is no IPTG, so bacteria survival depends on RBS strength. With strong RBS there will be more bacteria than with weak RBS.
The number of bacteria can be described as a parameter. In the model, number of bacteria in the laboratory is labeled as Z0. And in the environment there will be Z1 bacteria with strong RBS, Z2 with medium and Z3 with weak.Relation between numbers of bacteria can be defined as an ineiquality:
Z3 < Z2 < Z1 < Z0