Team:Vilnius-Lithuania/Measurement
Description
This year Vilnius iGEM team is also participating in the iGEM Measurement InterLab study. This is the second year that iGEM holds an InterLab study. This is one massive amazing experiment which involves multiple teams all around the world working for the same scientific goal.
This year the aim of this study is to see how different laboratories perform the same experiment and how results between different laboratories can differ or resemble each other. The ultimate goal of InterLab study is to develop new methods of interpreting data, which would take into account the differences that might arise whilst performing similar experiments in different conditions.
All teams had to construct three different GFP expressing devices and subsequently measure their fluorescence output. Received fluorescence data reflects the strength of gene (GFP) expression.
Three devices had to be constructed:
J23101, J23106 and J23117 parts are different promoters characteristic of different transcription levels. I13504 is ribosome binding sequence (RBS) + GFP + two terminators.
Experiments
Cloning
Cloning was performed using Biobrick Standard Assembly method. All cloning procedures were carried out in pSB1C3 vector.
Firstly, plasmids, containing three promoters and GFP gene, were transformed into E. coli DH5α strain. Bacteria, containing the plasmids, were grown overnight and then DNA extraction ensued. The extracted DNA was cut using EcoRI and SpeI restriction enzymes for the promoters, and with EcoRI and XbaI for plasmids, containing the GFP gene. Later, plasmids with incorporated GFP gene were purified using PCR cleaning kit and the promoters were extracted from gel using freeze and squeeze protocol. Then the promoters were ligated into vectors, containing GFP gene, ligation products were again transformed into E. coli DH5α strain. The validity of our constructs was tested by restriction mapping – using StyI restrictase.
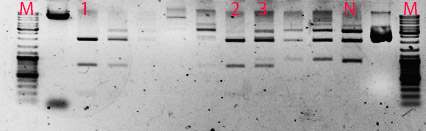
Growing
All three constructs were grown as three biological replicates. We used positive control – a GFP expressing device – BBa_I20270. We also used a negative control, which did not have the GFP gene in it – BBa_R0040. The positive and negative controls were grown in biological triplicates. All bacteria were grown in a LB medium overnight.
OD measurements
Optical density (OD) of bacteria grown overnight was measured and subsequently diluted to an OD600 value of 0.5.
Fluorescence measurements
After dilutions to OD600= 0.5, bacteria were measured for their fluorescence in a Jobin Ivon HORIBA FluoroMax-3 spectrofluorometer. The excitation was carried out at 500 nm and emission was recorded at 511 nm.
Results

Here we present the obtained fluorescence data. The histogram shows average fluorescence levels of bacterial populations with different investigated constructs. The chart also includes error bars.
Discussion
We see a huge difference in fluorescence between our positive and negative control. It means our experiment is working. The results show that both of bacteria with J23101 + I13504 and J23106 + I13504 constructs are characteristic of high GFP expression, thus the high fluorescence levels (bigger than the ones of the positive control). J23106 + I13504 constructs containing bacteria seem to fluoresce a little bit less than the J23101 + I13504 containing bacteria and we can see that J23117 + I13504 bacteria's fluorescence levels are a lot lower than the ones of the both other constructs, though higher than the ones of the negative control.
This data indicates that bacteria with different constructs are characteristic of different GFP expression levels. These levels should in turn represent different levels of transcription of the GFP gene, because only the promoters were different in three investigated constructs. In relation with these facts, we can conclude that J23101 is the strongest promoter, J23106 is a strong promoter and J23117 is a weak promoter in comparison to each other and the negative and positive controls.




