Difference between revisions of "Team:UCLA/Notebook/Honeybee Silk/14 July 2015"
(→Calculating Competency) |
|||
| Line 426: | Line 426: | ||
| − | =''' | + | ='''Ligation of SIlk + SpyCatcher into PSB1C3'''= |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
*3:1 insert to vector ratio. | *3:1 insert to vector ratio. | ||
{| class="wikitable" style="text-align:center; width:400px; height:200px;" | {| class="wikitable" style="text-align:center; width:400px; height:200px;" | ||
| Line 457: | Line 451: | ||
|} | |} | ||
*Program = 16C overnight and heat inactivate 10 min | *Program = 16C overnight and heat inactivate 10 min | ||
| + | |||
| + | |||
| + | ='''Calculating Competency'''= | ||
| + | *Calculating competency of cells prepared on 7/12. | ||
| + | *Equation = # colonies / ug DNA plated | ||
| + | *Using 2 ul of 50pg/ul puc19 vector to transform in to 50 ul | ||
| + | *Counted 300 colonies | ||
| + | *plated 1x10^-5 ug (1/10 of transformation reaction). | ||
| + | *Therefore tranformation efficiency = '''3 x 10^7''' cfu/ug DNA | ||
Latest revision as of 21:47, 24 August 2015
Contents
Miniprep
Samples 1 and 3 (inoculated) were miniprepped using the Zymo Plasmid Miniprep Kit.
OD
- 128.23 ng/uL
- 106.85 ng/uL
Both samples were sent in for sequencing.
PCR of Lac promoter/Silk/Spycatcher
- Using primer 3 and 4 that contain overhangs with the Biobrick prefix and suffix.
| Component | Volume (out of 50uL) |
|---|---|
| 5X Q5 Reaction Buffer | 10uL |
| 10mM dNTPS | 1uL |
| 10mM primer 3 | 2.5uL |
| 10mM primer 4 | 2.5uL |
| Template (Honeybee G-block) | 1uL |
| Q5 High Fidelity DNA Polymerase | 0.5uL |
| Nuclease Free Water | 32.5uL |
Program
| Step | Temperature | Time |
|---|---|---|
| Initial Denaturation | 98C | 30s |
| Cycles (x25) | 98C | 10s |
| Annealing | 72C | 20s |
| Extension | 72C | 30s |
| Final Extension | 72C | 2min |
| Hold | 12C | Hold |
Gel Visualization and Purification
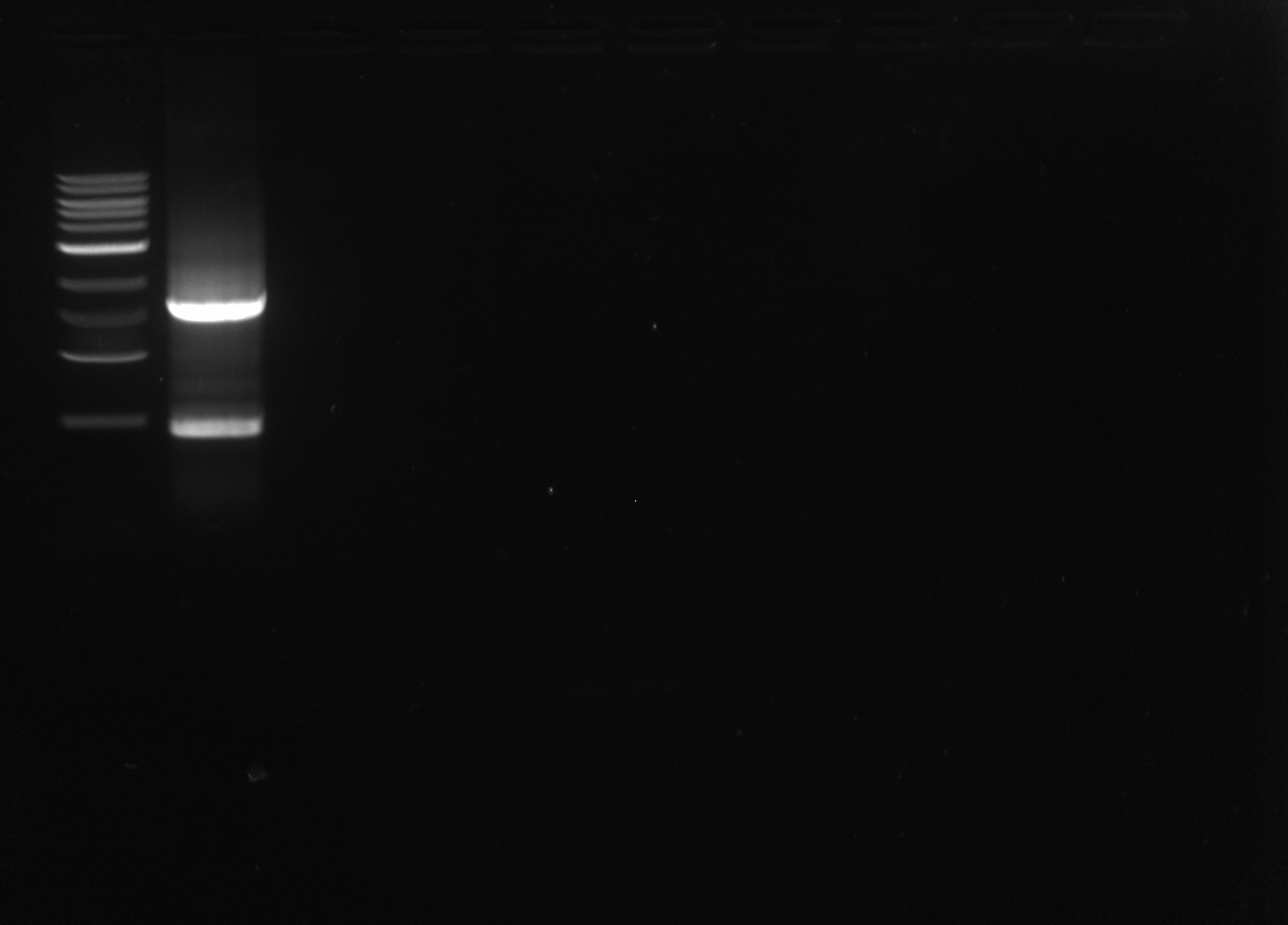
I ran the PCR product on a 1% agarose gel, expected size ~1600bp
Results: There was a band at the appropriate length and another band, probably the result of non-specific binding. The correct band was extracted and purified using the Qiagen Gel Purification kit.
OD: 88.12ng/uL
Double Digestion of Purified PCR Product
- The insert will be digested with EcoRI and Pst1 for preparation for ligation into the psb1c3 backbone (already digested with PstI and EcoRI).
| Component | Volume (out of 50uL) |
|---|---|
| NEB Buffer 2.1 | 5uL |
| EcoRI | 1uL |
| PstI | 1uL |
| Purified Insert | 9uL |
| Nuclease Free Water | 34uL |
Program
| Step | Temperature | Time |
|---|---|---|
| Incubation | 37C | 1 hour |
| Heat Inactivation | 80C | 15 min |
| Hold | 10C | Hold |
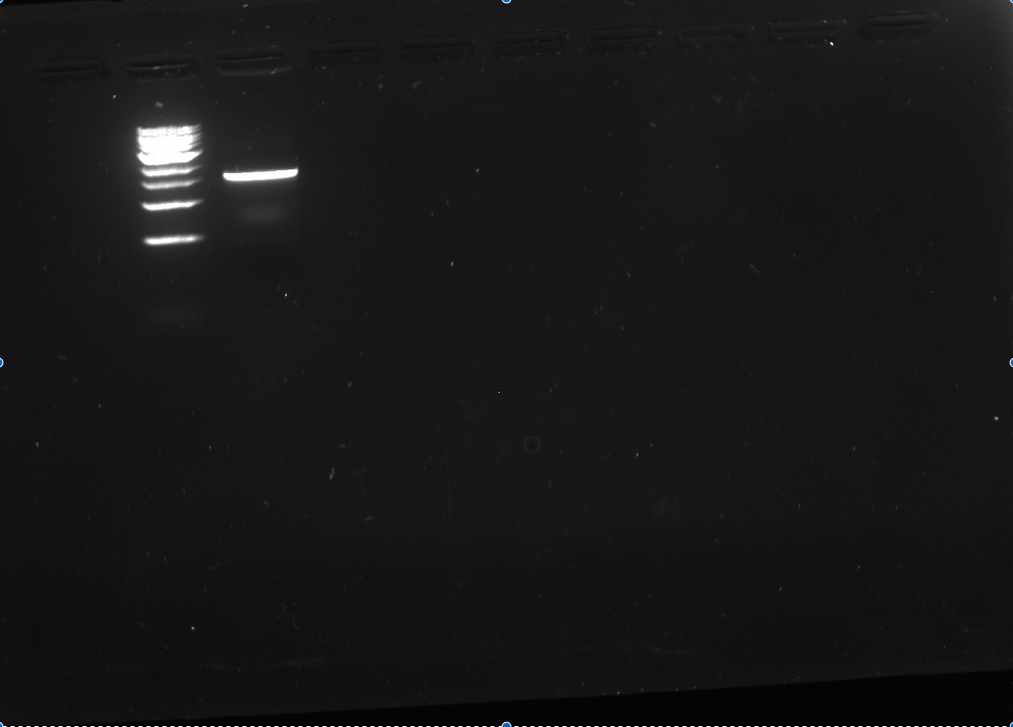
- Ran the digestion on a gel (all 50 ul) and compared to NEB 1 kb ladder. There was a nice clean band at the appropriate size.
Ligation of SIlk + SpyCatcher into PSB1C3
- 3:1 insert to vector ratio.
| Component | Volume (out of 50uL) |
|---|---|
| T4 Ligase Reaction Buffer | 2 ul |
| Backbone (psb1c3) | 1uL (50 ng) |
| Insert (silk + spyC) | 10ul (120 ng) |
| H20 | 6 ul |
| Ligase | 1uL |
- Program = 16C overnight and heat inactivate 10 min
Calculating Competency
- Calculating competency of cells prepared on 7/12.
- Equation = # colonies / ug DNA plated
- Using 2 ul of 50pg/ul puc19 vector to transform in to 50 ul
- Counted 300 colonies
- plated 1x10^-5 ug (1/10 of transformation reaction).
- Therefore tranformation efficiency = 3 x 10^7 cfu/ug DNA