Difference between revisions of "Team:Czech Republic/Notebook"
(→Module 3) |
(→Module 3) |
||
| (3 intermediate revisions by 3 users not shown) | |||
| Line 697: | Line 697: | ||
=== Module 3 === | === Module 3 === | ||
| − | + | The strength of the Anti-EpCAM scFv to EpCAM bond formation was examined. | |
| − | Yeast cells (EBY100) transformed with yeast display plasmid containing EpCAM and Anti-EpCAM scFv inserts were grown in induction medium for 48 hours ( | + | Yeast cells (EBY100) transformed with yeast display plasmid containing EpCAM and Anti-EpCAM scFv inserts were grown in induction medium for 48 hours (20°C). |
| − | After induction, cells were thoroughly vortexed to get rid of any | + | After induction, cells were thoroughly vortexed to get rid of any pre-formed clumps. |
| − | Samples containing only negative control (displaying only Anti-EpCAM scFv) and samples containing both of the yeast types (yeast cells displaying anti-EpCAM scFv and yeast cells displaying EpCAM), were incubated for 5 minutes in | + | Samples containing only the negative control (displaying only Anti-EpCAM scFv) and samples containing both of the yeast types (yeast cells displaying anti-EpCAM scFv and yeast cells displaying EpCAM), were incubated for 5 minutes in 30°C. |
| − | Solution of the medium and yeast cell aggregates was then qualitatively | + | Solution of the medium and yeast cell aggregates was then qualitatively transferred onto a microscope slide, covered with a coverslip and examined. |
| − | '''Summary: We successfully observed clump formations in examined | + | We also ran an assay with yeast cells containing plasmids for copper inducible production of mating factor/display of Anti-EpCAM scFv on the surface of the yeast cell and cells containing plasmids for pheromone inducible production of GFP/display of EpCAM. We mixed these cells directly on the microreader (1:1) and induced them with the copper sulfate solution. We also had negative controls, which were cells transformed with the same plasmids except for the yeast display plasmids, therefore they could not bind each other. |
| + | |||
| + | '''Summary: We successfully observed clump formations in the examined samples. Fluorescence in the mixed culture was higher than in the cultures containing only one type of yeast.''' | ||
=== Module 5 === | === Module 5 === | ||
| Line 710: | Line 712: | ||
Signal transduction experiments in microfluidics: synthetic MATa producing alpha-pheromone + MATa pFUS1-GFP - done by Martin Cienciala. | Signal transduction experiments in microfluidics: synthetic MATa producing alpha-pheromone + MATa pFUS1-GFP - done by Martin Cienciala. | ||
| + | |||
| + | Yeast-induced blood agglutination on-chip - done by Martin Cienciala. | ||
---- | ---- | ||
Latest revision as of 03:19, 19 September 2015
Notebook
Contents
May
Week 1 (4-10)
Module 1
Extraction of MATx genome - used strain 7283: MATx ura3-52 Gal+ (Genome extraction)
PCR amplification of all 6 parts of the final MATx intergration plasmid (PCR):
- MATalpha2 (template 7283 genome)
- CYC1v3 (template gBlock)
- pRS406 (template miniprep)
- STE12 w/ 3'UTR (template 7283 genome)
- pTv3 double promoter (template gBlock)
- MATalpha1 (template 7283 genome).
Gibson assembly (Gibson assembly)
- 2-round Gibson: first round with fragments 2-5, than MATalpha2 and MATalpha1 added
- 1-round Gibson: all parts together
Transformation of both Gibson assembly reactions into e. coli. Colony PCR showed unsuccessful Gibson assembly. (Transformation, Colony PCR, Gel electrophoresis)
Gibson assembly (Gibson assembly)
- reaction: parts 1, 2, 6
- reaction: parts 3, 4, 5
PCR amplification of fragments from Gibson reactions. Gel showed that Gibson 6,1,2 has the correct size. (PCR, Gel electrophoresis)
Gibson assembly 6,1,2+4+5 (Gibson assembly)
PCR amplification of fragments from Gibson reactions. Gel showed that Gibson 6,1,2+4+5 has the correct size. (PCR, Gel electrophoresis)
Gibson assembly 4,5,6,1,2 + 3 (vector). Gibson reaction was transformed into e-coli. Colony PCR showed unsuccessful Gibson assembly. (Gibson assembly, Transformation, Colony PCR, Gel electrophoresis)
Hybrid - we need to have a reporter plasmid for measure the function of hybrid plasmid pH30 (trp auxotrophy) (gift form prf. Fields).
PCR of GFP for hybrid, as a template was a plasmid from the laboratory depository. ((PCR)
Cloning (restriction, ligation, transformation) the PCR GFP construct into pRSII416 vector. (Restriction digest, Ligation, Transformation)
Summary: We successfully extracted parts of MATx genome and connected 5 parts of synthetic MATx via Gibson assembly. We successfully construct GFP on pRSII416.
Module 2
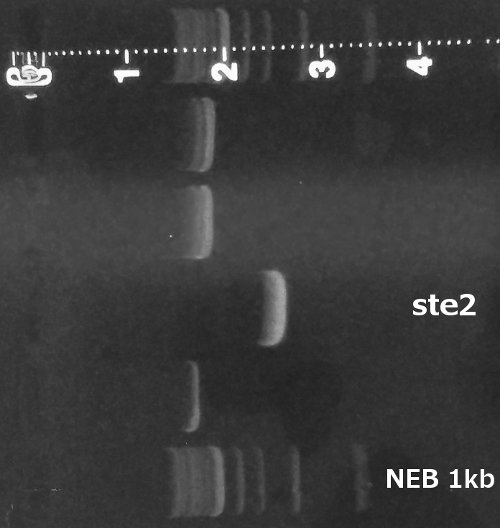
PCR of the ste2 gene from the genome of Saccharomyces cerevisiae 7284 strain (Genome extraction, PCR, Gel electrophoresis).
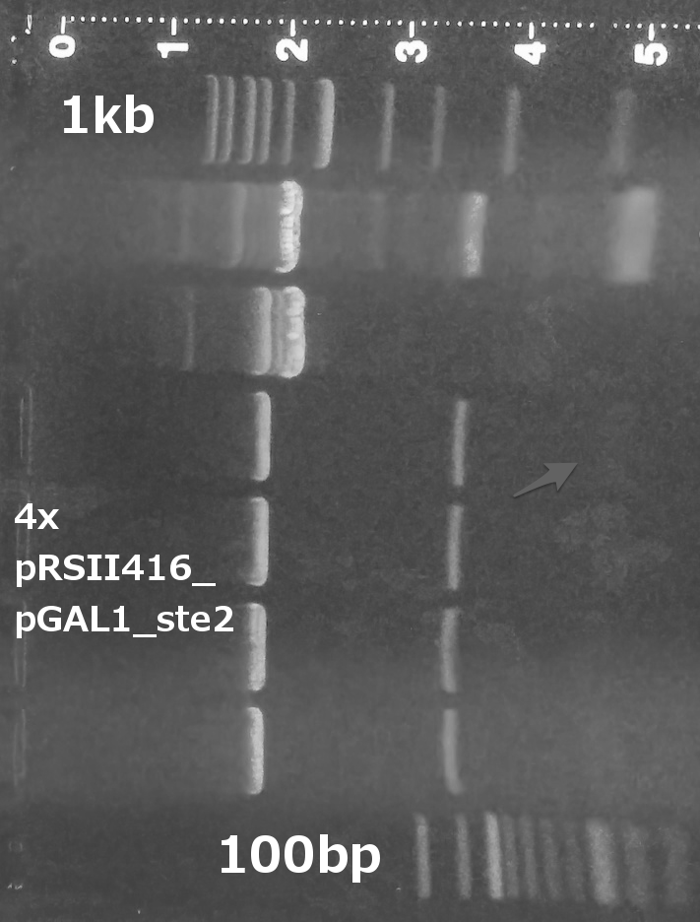
Cloning of the amplified ste2 into pRSII416 vector (Restriction digest, Ligation, Transformation).
Verification of the pRSII416_ste2 construct (Miniprep, Restriction digest, Gel electrophoresis).
Summary: We successfully obtained ste2 gene from the yeast genome and cloned it into pRSII416 vector.
Module 3
Restriction testing of the pCT302 yeast display plasmid.
Auxotrophy testing of the yeast display yeast strain - EBY100.
Summary: We tested the EBY100 strain and pCT302 plasmid we recieved as a gift. Tests proved that we are ready for cloning of the streptavidin open reading frame.
Week 2 (11-17)
Module 1
Gibson assembly 4,5,6,1,2 + 3 (vector). Transformation into e. coli. 6 colonies obtained. Minipreps and restriction digest of all colonies. (Gibson assembly, Transformation, Miniprep, Restriction digest, Gel electrophoresis)
5 from 6 colonies that grow from Gibson assembly reaction show bands with correct sizes - MATX integration plasmid has been assembled successfully.
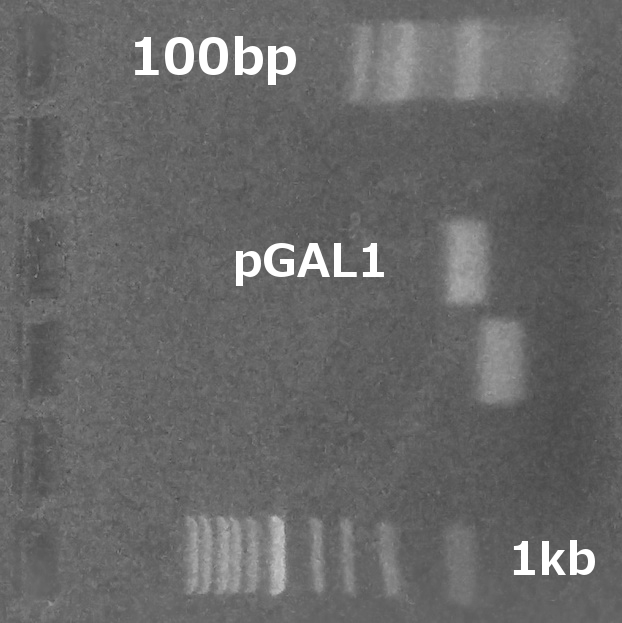
Cloning (restriction, ligation, transformation) pGAL1 promoter (obtained from the Registry, part BBa_J63006) into plasmid GFP on pRSII416. Sending the construct to sequencing. (Restriction digest, Ligation, Transformation)
Summary: We successfully assembled synthetic MATx integration plasmid, sequencing will follow. The sequencing results of pGAL1 promoter- GFP-pRSII416 show no GFP on the plasmid, but the presence of pGAL1 was verified.
Module 2
PCR of the incomplete MF(ALPHA)1 locus (see the Design page for details) from the genome of Saccharomyces cerevisiae 7283 strain (Genome extraction, PCR, Gel electrophoresis).
Band-stab PCR of the amplified incomplete MF(ALPHA)1 locus in order to add the actual pheromone and a stop codon.
Cloning of the obtained pheromone into pRSII416 vector (Restriction digest, Ligation, Transformation).
Verification of the pRSII416_SC_alpha construct (Miniprep, Restriction digest, Gel electrophoresis).
Summary: We successfully obtained mating pheromone from Saccharomyces cerevisiae with its secretion tag and cloned it into pRSII416 vector. We also prepared pCUP1 promoter for the cloning upstream of this pheromone.
Module 3
PCR amplification of the streptavidin gBlock with M13 primers (PCR).
Restriction digest of the PCR product and pCT302 vector. Digested products were gel purified and ligated. Transformation of DH5alpha bacteria cells with ligation product (Restriction digest, Ligation).
Summary: Streptavidin ORF was successfully cloned into yeast display vector pCT302.
Week 3 (18-24)
Module 1
Sequencing of synthetic MATx insert.
New PCR of GFP and new cloning (restriction, ligation, transformation) was done this time into previous plasmid pGAL1 on pRSII416. (PCR, Restriction digest, Ligation, Transformation)
Plate results: colonies with the construct pGAL1_GFP on pRSII416.
Summary: Sequencing confirmed successful assemble of synthetic MATx. We obtained the bacterial colonies with the construct pGAL1_GFP on pRSII416. Minipreps will be done next week.
Module 2
PCR amplification of pGAL1 promoter from the genome of Saccharomyces cerevisiae 7284 strain (PCR, Gel electrophoresis).
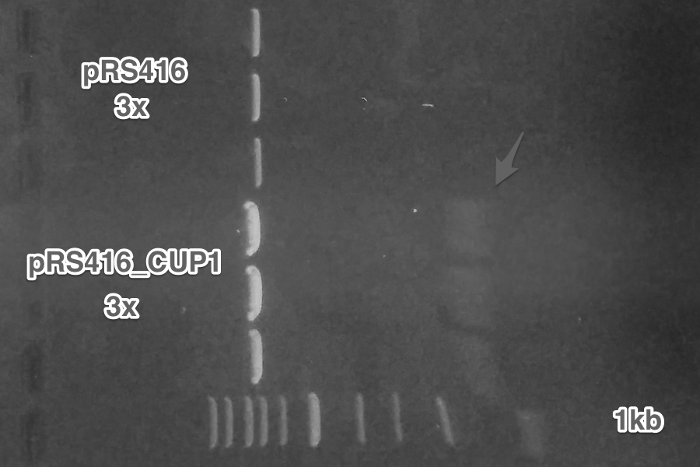
Cloning of the amplified pCUP1 from Week 2 into pRSII416_SC_alpha plasmid (Restriction digest, Ligation, Transformation).
Verification of the pRSII416_pCUP1_SC_alpha construct (Miniprep, Restriction digest, Gel electrophoresis).
Summary: We have successfully obtained plasmid with a mating pheromone under the control of pCUP1 promoter and prepared pGAL1 promoter for the control of ste2 gene.
Module 3
Miniprep from selected transformed colonies and restriction digest of isolated plasmids (Restriction digest).
Gel verification and sequencing (Gel electrophoresis).
Summary: Transformed colonies had correct vectors. Sequencing data showed us that streptavidin gene is inserted in frame and is without any mutations.
Week 4 (25-31)
Module 1
Extraction of MATa locus from genome of MATa strain in order to obtain precise sequence of MATa locus. PCR of MATa locus for sequencing. Both steps 3x repeated. Gel always showed unsuccessful extraction of MATa strain. (Genome extraction, PCR, Gel electrophoresis)
Miniprep from selected colonies and the restriction for verification of the insert pGAL1_GFP. (Miniprep, Restriction digest, Gel electrophoresis).
Summary: We successfully constructed the plasmid pGAL1_GFP on pRSII416. We unsuccessfully tried to extract MATa locus from genome.
Module 2
Summary: We successfully cloned pGAL1 promoter to the pRSII416_ste2 plasmid and verified the final constructs using gel electrophoresis.
June
Week 5 (1-07)
Module 1
PCR of MATa locus for sequencing. Gel showed weak band with right length. Another PCR was run to get more amplification. Samples were sent for sequencing. (Genome extraction, PCR, Gel electrophoresis)
Transformation of the pGAL1_GFP on pRSII416 into yeast strain 7284 MATa. At the end of the week the assay was running. (Transformation)
Summary: We successfully extract MATa locus from genome. We obtained the yeast transformants with the pGAL1_GFP on pRSII416. The assay shows no fluorescence.
Module 2
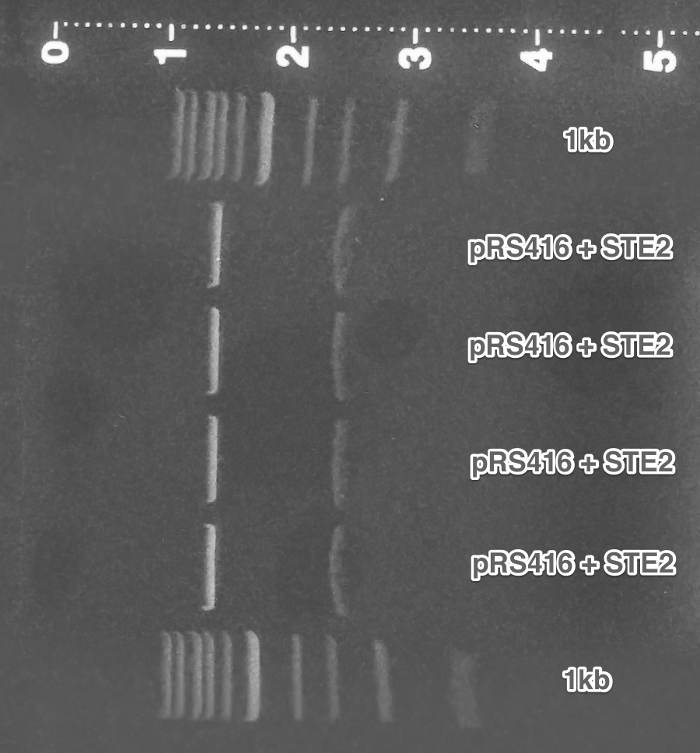
Verification of the pRSII416_pGAL1_ste2 and pRSII416_pCUP1_SC_alpha plasmids. The results can be accessed here.
Summary: The function of the two constructed plasmids so far has been verified.
Week 6 (08-14)
Module 1
Repetition of the assay but this time we used the minimal drop in medium instead of the rich one.
Summary: The assay shows no fluorescence
Module 2
Preparation of the gBlocks to be ordered (STE2 receptors from different yeast strains).
Summary: We have designed five different gBlocks and ordered them using the promotion from the IDT.
Week 7 (15-21)
Module 1
PCR amplification of pRSII403 for Gibson assembly. Unsuccessful. Repeated. Gel showed weak band of the correct size. (PCR, Gel electrophoresis)
Gibson assembly of STE12+pRSII403. PCR amplification of STE12-pRSII403 fragment. There were very weak bands of the correct size 6.5kb. (Gibson assembly, PCR, Gel electrophoresis)
G-blocks yG-MATa_1 and yG-MATa_2 were ordered.
PCR of GFP this time with primers whose added the Kozak sequence. The template was a plasmid from the laboratory depository, the same as previously. ((PCR)
Cloning (restriction, ligation, transformation, colonies, miniprep, restriction, gel) the PCR GFPK (GFP with Kozak sequence) construct into plasmid with pGAL1 on pRSII416 vector. (Restriction digest, Ligation, Transformation, Miniprep, Restriction digest, Gel electrophoresis))
Summary: We successfully construct GFPK on pRSII416. We ordered G-blocks for synthetic MATa assembly.
Module 2
Primers design and order for the construction of different pheromones (for the site-directed mutagenesis).
Summary: All primers needed for a construction of plasmids with different pheromones were designed and ordered.
Week 8 (22-28)
Module 2
Restriction of the gBlocks ordered in Week 6 and of the pRSII416_pGAL1_ste2 plasmid (Restriction digest).
Summary: We have prepared vector with pGAL1 promoter for the different ste2 genes. All gBlocks we recieved were prepared for the ligation to these vectors.
Week 9 (29-05)
Module 1
G-blocks yG_MATa_1 and yG_MATa_2 arrived. PCR amplification of MATa_1 and MATa_2. Gel verification showed successful amplification of MATa_1 - strong band, unsuccessful amplification of MATa_2.(PCR, Gel electrophoresis)
Repeated PCR Amplification of MATa_1 and MATa_2. Gel verification showed weak bands and possible contamination. (PCR, Gel electrophoresis)
Repeated PCR Amplification of MATa_1 and MATa_2.Gel verification showed strong correct band of MATa_1 and weak correct band of MATa_2.(PCR, Gel electrophoresis)
We proceed the transformation of the pGAL1_GFP on pRSII416 into yeast strain 6193 MATa and the transformation of the pGAL1_GFP on pRSII416 and pH30 (gift from prof. Fields) into 6193 MATa. ((Transformation))
Summary: We recieved yG_MATa_1 and yG_MATa_2 G-blocks and successfully amplified them. We obtained the yeast transformants with the pGAL1_GFP on pRSII416 and with two plasmids - with the pGAL1_GFP on pRSII416 and hybrid plasmid.
Module 2
Ligation of the digested gBlocks from Week 8 to the pRSII416_pGAL1 vectors, transformations to bacteria, patches, minipreps and verification by restriction digest and gel electrophoresis (Ligation, Transformation, Gel electrophoresis).
At least one of the three colonies tested showed the correct length of the insert.
Summary: We obtained plasmids with different STE2 receptors.
Module 5
PDMS molding (40μm high silicon masters) - done by Vaclav Pelisek.
July
Week 10 (06-12)
Module 1
Gibson assembly of MATa_1 + MATa_2 + STE12 insert and MATa_1 + MATa_2. PCR amplification of Gibson assembly. Gel verification - empty gel. (Gibson assembly, PCR, Gel electrophoresis)
PCR amplification of STE12 3UTR and STE12 3UTR from genome. Gel verification showed strong correct bands. Purified STE12 were both correct and usable for Gibson assembly. (PCR, Gel electrophoresis)
PCR amplification of pRSII403. Gel verification of pRSII403 - clean and strong bands in all rows. Vector ready to be used in Gibson Assembly. (PCR, Gel electrophoresis)
Gibson assembly of MATa Insert (MATa_1 + MATa_2 + STE12). PCR amplification of MATa insert. Gel verification of MATa PCR - extremely weak (barely visible bands). Repeated PCR amplification of MATa insert. Gel verification - empty gel. (Gibson assembly, PCR, Gel electrophoresis)
PCR amplification of MATa insert by parts - tried to amplify MATa_1+MATa_2, MATa_2 + STE12. Gel verification - two extremely weak incorrect bands. GA did not connect even two parts. (PCR, Gel electrophoresis)
Gibson Assembly of MATa (MATa_1 + MATa_2 + STE12 + pRSII403). Transformation of MATa Gibson Assembly into E.Coli. (Gibson assembly, Transformation)
Hybrid and pGAl_GFP reporter plasmid assay.
Summary: We successfully amplified pRSII403, STE12 3UTR and STE12 3UTR from genome. The assay was successful. We obtained expected results.Gibson Assembly of MATa was not successful.
Module 2
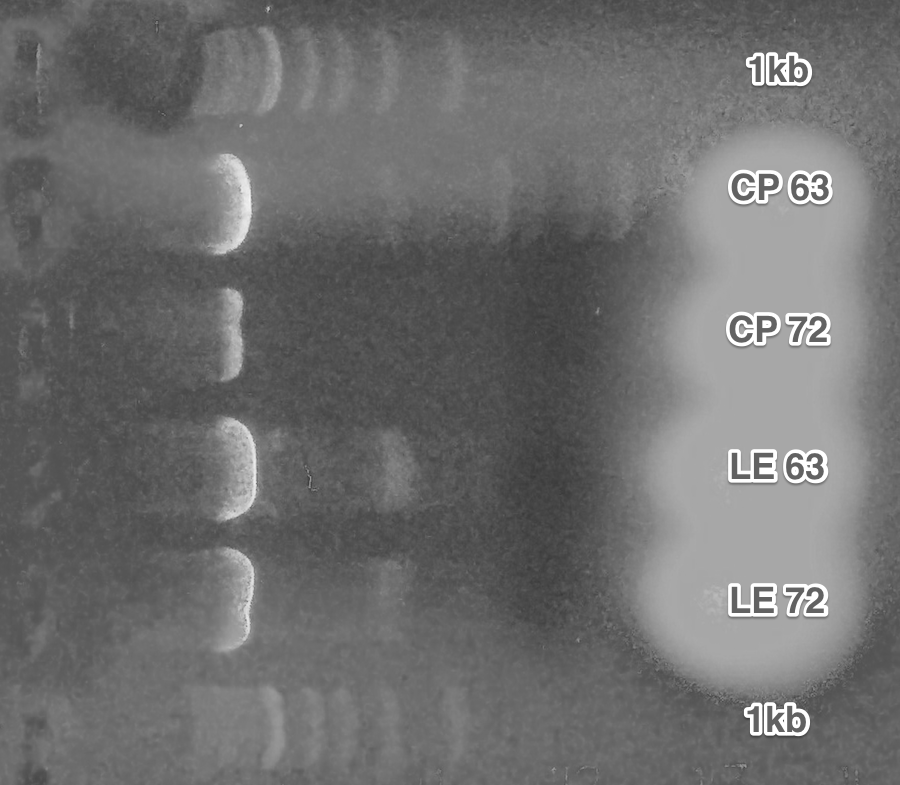
Creation of the plasmids with different alpha-factor pheromones (Site-directed mutagenesis).
As can be seen on the picture, each pheromone SDM had to be performed with different annealing temperature.
Transformations, patches, minipreps, and verification of the obtained plasmids (Miniprep, Restriction digest, Gel electrophoresis).
All screened plasmids with the exception of LE6 showed the desired length of the insert.
Summary: Plasmids with different pheromones were successfully obtained.
Week 11 (13-19)
Module 1
Plate results of transformation of MATa form last week - numerous colonies. Minipreps of MATa GA and verificaiton restrictions. Gel verification - only vectors were linked by the GA, no inserts. (Miniprep, Restriction digest, Gel electrophoresis)
Hybrid and pGAl_GFP reporter plasmid assay in the rich medium.
Summary: The assay was successful. We obtained expected results. Gibson Assembly of MATa was not successful.
Module 2
Transformations of plasmids with pheromones and receptors to the S. cerevisiae a cells (Δbar1 and 7284). (Transformation (yeast))
Week 12 (20-26)
Module 1
Gibson assembly of MATa (MATa_1 + MATa_2 + STE12). PCR amplificaiton of GA. Gel verification - mutliple bands of incorrect size. (Gibson assembly, PCR, Gel electrophoresis)
Gibson assembly of pRSII403+MATa_1. PCR amplification of pRSII403+MATa_1. Gel verification - unsuccessful GA. (Gibson assembly, PCR, Gel electrophoresis)
A-specific CYC1 promoter amplified from yG_MATa_2. (PCR)
Minipreps of pRS416-pSTE2-GFP. PCR of pRS416-GFP (Minipreps of pRS416-pSTE2-GFP were used as templates for PCR.) Gel verification - bands of incorrect sizes only. (Miniprep, PCR, Gel electrophoresis)
Restriction of reporter plasmids: (Restriction digest)
- pFUS1 (XhoI, HindIII)
- pSTE5 (XhoI, HindIII)
- pTv3 (XhoI, HindIII)
- asCYC1 (XhoI, EcoRI)
- pSTE2-GFP (XhoI, HindIII)
- pSTE2-GFP (XhoI, EcoRI)
Ligation of pRS416-GFP with pFUS1, pSTE5, pTv3, asCYC1. Transformation into E. coli. (Ligation, Transformation)
PCR overlap extension:
- MATa insert (MATa_1, MATa_2, STE123UTR)
- MATa_1 + MATa_2
- MATa_2 + STE123UTR
Gel verification: unsuccessful
Summary: All reporter plasmids were prepared and transformed into E. coli. Verifications will be done next week. Gibson Assembly of MATa was not successful.
Week 13 (27-02)
Module 1
Minipreps of reporter plasmids from last week. Restriction of minipreps - XhoI+HindIII. Verification gel - only asCYC1 and pTv3 hadthe right bands. 3 more colonies of both pFUS1 and pSTE5 were streaked for patches. (Miniprep, Restriction digest, Gel electrophoresis)
Minipreps of pFUS1 and pSTE5. Restriction. Verification gel - one correct pFUS1. (Miniprep, Restriction digest, Gel electrophoresis)
Ligation of pSTE5 into pRS416-GFP. Transformation into E. coli. (Ligation,Transformation)
Minipreps of pSTE5_pRS416-GFP. Restriction XhoI+SpeI. Verification gel - pSTE5-GFP confirmed. (Miniprep, Restriction digest, Gel electrophoresis)
PCR overlap extension of MATa_1 + MATa_2. Gel verification - bands of both corrent and incorrect length. Band of the correct size was cut-out and cleaned. (Gel electrophoresis)
PCR overlap extension MATa_1_MATa_2 + STE123UTR. Gel verification - unsuccessful PCR overlap extension. (Gel electrophoresis)
Sequencing results of MATa_1_MATa_2 fragment - alignment shows 2 errors in TetR sequence.
Summary: Gel verification of pFUS1, pSTE5, pTv3 and asCYC1 reporter plasmids showed successful assembly. Sequencing will follow. PCR overlap extension of MATa_1 + MATa_2 was not successful.
Module 2
Orthogonality tests using the growth arrest approach. The results can be found on our project page together with the protocol.
Summary: We were able to identify two orthogonal signals and their receptors - from S. cerevisiae and C. parapsilosis.
August
Week 14 (03-09)
Module 1
We decided to assemble MATx locus via ligation.
Restriction: (Restriction digest)
- pRSII406 with SalI+HindIII - for cloning STE123UTR for both MATa and MATx
- pSB1C3 with XbaI+SpeI (for biobricks)
PCR (template: genome): (PCR)
- STE12 3UTR - For both MATa and MATx gBlocks and MATa LIGATED
- pTv3pTv3 - For Insert MATx w/Kozaks
- MATa1_MATa2 - For Insert MATa LIGATED
Gel verification - successful (Gel electrophoresis)
PCR: (PCR)
- Cyc1v3-pRSII406-STE12 3 UTR - For Insert MATx w/Kozaks
- vector for Cyc1v3 - For pRS416-CYC1v3-GFP
Gel verification - vector for CTC1v3 correct (Gel electrophoresis)
PCR: (PCR)
- Cyc1v3-pRSII406-STE12 3 UTR - For Insert MATx w/Kozaks
- MATa1_MATa2 - For Insert MATa LIGATED
Gel verification - successful (Gel electrophoresis)
Restriction: (Restriction digest)
- pADH1 (XhoI+HindIII)
- CYC1v3 (EcoRI+HindIII)
- STE12 3UTR (HindIII+SalI)
- vector for Cyc1v3 (EcoRI+HindIII)
Ligation of (Ligation)
- pADH1 into pRS416-GFP
- CYC1v3 into pRS416-GFP
- STE123UTR into pRSII406
Transformation into E. coli (Transformation)
PCR: (PCR)
- Cyc1v3-pRSII406-STE12 3 UTR - For Insert MATx w/Kozaks
- MATa1_MATa2 - For Insert MATa LIGATED
Verification gel - correct bands were cut-out from the gel and cleaned (Gel electrophoresis)
Transformations into 7284: pSTE2, pSTE5, pFUS1, pTV3, asCYC1 (Transformation)
Minipreps: (Miniprep)
- pRS416-pADH1-GFP (2X)
- pRS416-CYC1v3-GFP (2X)
- pRSII406-STE123UTR (2X)
Restriction of minipreps. Verification gel - pADH1 has the correct bands. One colony that left on plate with CYC1v3 tranformants was streaked for patch and an electroporation with 5 ul of ligation will be repeated. 6 colonies from pRSII406-STE123UTR transformants were streaked for patches. (Restriction digest, Gel electrophoresis)
Yeast Transformations into 10150: pSTE2, pSTE5, pFUS1, pTV3, asCYC1, pADH1 (Transformation)
Summary: We decided to assemble MATx locus via ligation. Reporter plasmids were transformed into yeasts. PCR for MATx ligation was prepared. pADH1 reporter plasmid was assembled
Module 3
Transformation of the EBY100 yeast cells with the streptavidin yeast display plasmid (Transformation).
Growing and induction of the yeast transformants in 20 °C and 30 °C. Induction in 20 °C was used because streptavidin monomers tend to fold incorrectly in higher temperatures.
Microfluidics experiments examined functionality of the displayed streptavidin. Microfluidics chips covered with biotinylated gelatin were used in our case.
Summary: Transformed yeast cells were not able to bind to the biotinylated surface. This could be due to a several reasons. Firstly, streptavidin monomers could be folded incorrectly. Secondly, functional streptavdin unit composes of four streptavidin monomers, there is only a little chance that four of the displayed monomers will be in the correct conformational positions to form functional streptavidin unit. Thirdly, there might not be enough of biotin molecules on the microfluidic chips for mediating the cell to chip connection. We also decided that expressing both Aga1 and Aga2 with constitutive promoter could be a better idea, as yeast with plasmids using galactose promoters need to grow in a medium containing sacharose/galactose ergo grow slower.
Week 15 (10-16)
Module 1
Gibson assembly MATx. Transformation of MATx gibson into E. coli. No colonies. (Gibson assembly, Transformation)
PCR of asCYC1, pTv3. (PCR)
Minipreps: (Miniprep)
- pCYC1v3
- pRSII406 + STE123UTR
Verification restriction. Gel verification - no CYC1v3 and pRSII406-STE123UTR (Restriction digest, Gel electrophoresis)
PCR of STE12 3UTR - for cloning into pRSII406 (PCR)
Ligation and transformation into E. coli: (Ligation, Transformation)
- pRSII406 + STE12
- pRSII416 + STE12
- pRSII416-GFP + FUS1
New G-blocks yG_MATa, yG_MATx_1, yG_MATx_2 arrived.
MAT knockout PCR. Verification gel - single band of the correct size (181bp). (PCR, Gel electrophoresis)
Restriction of MAT knockout. Purification from gel of MATa biobrick insert, MAT knockout, pSB1C3. (Restriction digest, Gel electrophoresis)
Colonies on plates of pRSII406 + STE12 and pRSII416 + STE12. Colony PCR of pRSII406 + STE12 3'UTR and pRSII416 + STE12 3'UTR. Verification gel - unsuccessful. (Colony PCR, Gel electrophoresis)
Repeated ligation of pRSII406 + STE12 and pRSII416 + STE12. Colonies on plates of pRSII406 + STE12 and pRSII416 + STE12. (Ligation, Transformation)
Minipreps. Verification reastriction and gel - unsuccessful. (Miniprep, Restriction digest, Gel electrophoresis)
Ligation of MATa and MAT knockout into pSB1C3. Transformation into E. coli. Transformation of pFUS1-GFP-pRSII416 ligation. (Ligation, Transformation)
Summary: We recieved G-blocks for MATx assembly via ligation. The ligation of STE12 and vectors was unsuccessful. We transformed ligations of biobricks. Results will be next week.
Module 2
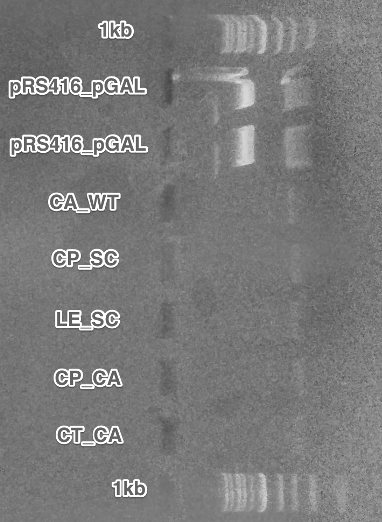
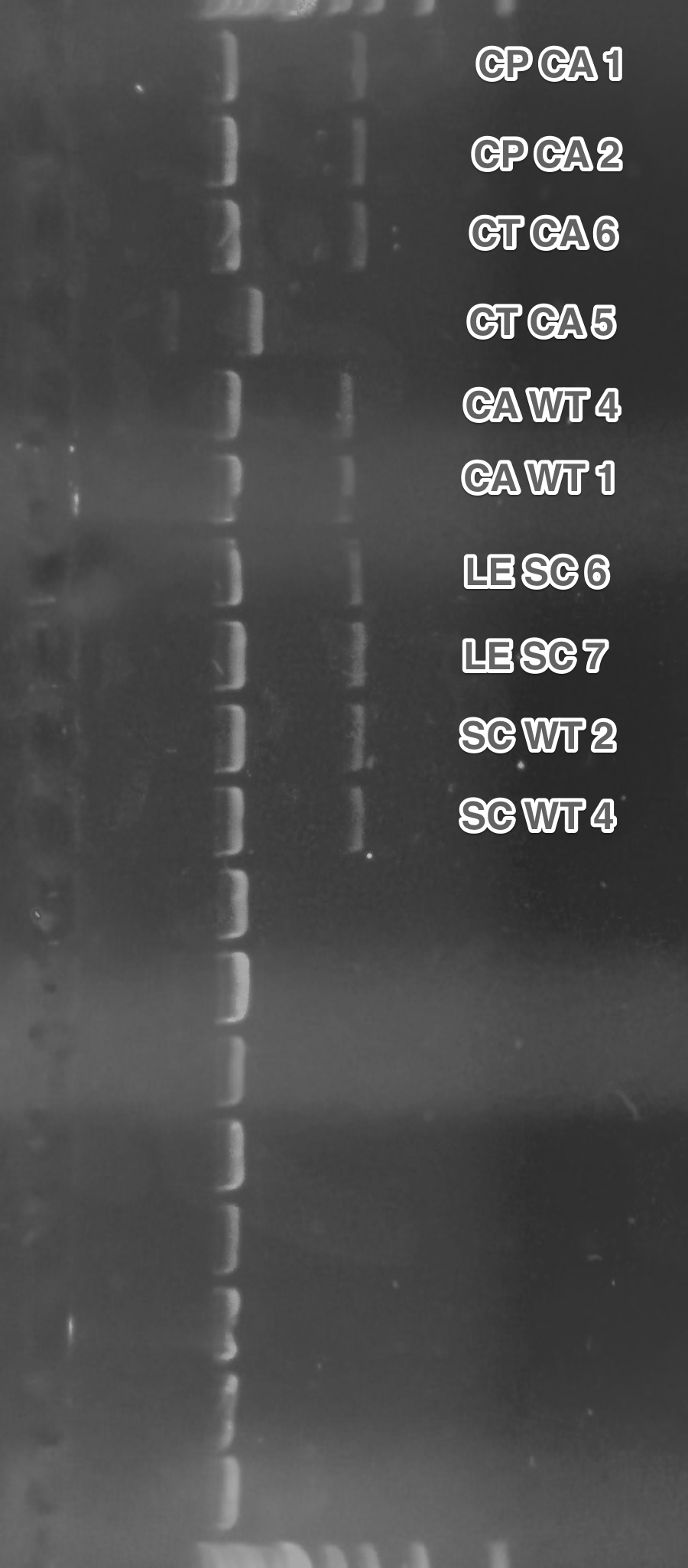
Replacement of the pGAL1 promoters on the receptors plasmids with pADH1 (Restriction digest, Gel electrophoresis,Ligation,Transformation).
Except for the CT CA 5, all promoters were successfully replaced.
Summary: We have obtained receptors under the control of constitutive pADH1 promoter. This will simplify our future experiments, since we won't need to grow transformed cells in a special medium.
Module 3
Adding of the restriction sites at the end of EpCAM gBlock. One reaction added restriction sites needed for cloning into yeast display plasmid, the other added restriction sites needed for cloning into pRSII plasmid needed for secrection of EpCAM protein (PCR).
Purification of the PCR products and subsequent restriction digest with according restriction enzymes. gBlocks encoding Anti-cMyc scFv, Anti-EpCAM scFv and Anti-HuA scFv were also digested, but only for the use in yeast display plasmids (Restriction digest).
Gel purified inserts were ligated either with the yeast display plasmid (pCT3O2) or the secretion plasmid - only EpCAM ORF (pRSII 416 with inducible CUP1 promoter and pheromone signalling sequence). Ligation products were transformed into DH5alpha bacteria cells (Ligation, Transformation).
Miniprep of transformants and restriction digest testing (Restriction digest).
Summary: We were unable to obtain any usable transformants with the secretion plasmid, however this project was canceled because of more pressing issues. Sequencing data showed that inserts in the yeast display plasmids are in frame and without any mutations.
Week 16 (17-23)
Module 1
Plate results:
- no colonies of pSB1C3 + MATa b.b ligation, pSB1C3 + MATa k/o
- colonies of pFUS1-GFP-416
Minipreps of pFUS1-GFP-416. Restriction. Gel verification - correct bands. (Miniprep, Restriction digest, Gel electrophoresis)
Restrictions of pRSII406 vector (HindIII+PstI, PstI+SpeI, HindIII+PstI). Gel verification and clean up - successful. (Restriction digest, Gel electrophoresis)
Ligation of MATx1, MATx2, MATx1 + MATx2, MATa into pRSII406. Gibson assembly of MATx. Transformation into E. coli. (Ligation, Gibson assembly, Transformation)
Colonies of MATx1, MATx2, MATx1 + MATx2, MATa in pRSII406. Minipreps of pRSII406-MATa, pRSII406-MATx1 + MATx2. Restriction and verification gel - correct. Synthetic MATx locus was successfully assembled. (Miniprep, Restriction digest, Gel electrophoresis)
Overnights for yeast transforms: 7283, 7284, 10150, 4741 delta Sst2 (Transformation)
Overnight ligation of pRSII406 S+H and STE123'UTR. Transformation into E. coli. (Ligation, Transformation)
Minipreps of pRSII406-STE12. Restriction and verification gel - all samples negative. (Miniprep, Restriction digest, Gel electrophoresis)
PCR of STE12 3'UTR. Restrictions of pRSII406. Gel verification - correct. (PCR, Restriction digest, Gel electrophoresis)
Yeast transformations of reporter plasmids pFUS1, pasCYC, pTv3, pSTE2, pSTE5 into 7284a, 10150 a/x and 7283x. (Transformation)
Restriction of pFUS1-GFP-416. Restriction of pRSII413. Gel verification - correct. (Restriction digest, Gel electrophoresis)
Ligation of pFUS1-GFP-413. Transformation into E. coli. No colonies. (Ligation, Transformation)
Ligation and transformation into E. coli: (Ligation, Transformation)
- pRSII406 (E+Sal) + yG_MATa + STE12 3'UTR (1:3:3) ligation
- pRSII406 (Sal + Spe) + yG_MATx1 + yG_MATX2 + STE12 3'UTR (1:3:3:3) ligation
- pRSII406-yG_MATa (HindIII + SalI) + STE12 3'UTR (1:6) ligation
- pRSII406-yG_MATx1-yG_MATx2 (HindIII + SalI) + STE12 3'UTR (1:6) ligation
Plate results:
- no colonies: pRSII406 + yG_MATa + STE12 3'UTR, pRSII406 + yG_MATx1 + yG_MATX2 + STE12 3'UTR
- colonies: pRSII406-yG_MATa + STE12 3'UTR, pRSII406-yG_MATx1-yG_MATx2 + STE12 3'UTR
Minipreps. Restriction. Verification gel - unsuccessful. (Miniprep,Restriction digest, Gel electrophoresis)
Phosphatase treatment on: RSII406-yG_MATa and pRSII406-yG_MATx1-yG_MATx2. Ligation after phosphatase treatment: (Ligation, Transformation)
- pRSII406-yG_MATa + STE12 3'UTR
- pRSII406-yG_MATx1-yG_MATx2 + STE12 3'UTR
- pRSII413 + pFUS1-GFP
Transformation into E. coli
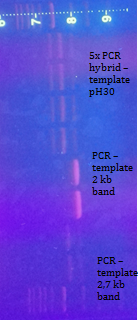
PCR of hybrid as a template was the plasmid pH30. The used primers were not original for this sequence. Done just for verification of annealing parts. (PCRGel electrophoresis)
Summary: We successfully assembled synthetic MATx locus. We successfully transformed reporter plasmids into yeasts. We were not able to clone STE12 3'UTR into pRSII406-yG_MATa or pRSII406-yG_MATx1-yG_MATx2. We obtained a band of proper length for hybrid transcriptional factor.
Module 2
Transfer of the receptors - to change the selectable marker to leu (Restriction digest, Ligation).
Summary: With all the receptors on the leu plasmid, we will be able to transform these plasmids together with the pFUS1_GFP receptor. This will allow us to test the orthogonality of receptors using fluorescence as the output.
Module 3
PCR of the pTV3 promoter and CYC1 transcription terminator (PCR).
Restricted PCR products were gel purified and ligated. Ligation product was PCR amplified with primers used in the preceding PCR reaction (5' primer for pTV3 promoter, 3' primer for CYC1 transcription factor) (PCR, Restriction digest, Ligation).
PCR product (expression cassette) was restricted for use in pRSII 413 and pRSII 415 plasmids (Restriction digest).
Ligation with given vectors and transformation of DH5aplha bacteria cells (Ligation, Transformation).
Summary: We were not successfull in creating the universal expression vectors (pRSII 413/415 + expression cassette).
Module 5
Bonding of PDMS to glass (6 microfluidic chips encapsulated on two glass slides) - done by Vaclav Pelisek.
Week 17 (24-30)
Module 1
Restrictions of pFUS-GFP-416 and pRSII413. Gel verificaiton and clean up of pFUS1-GFP and pRSII413 - Bands of the correct sizes in all lanes, excised and cleaned. Ligation of pRSII413+pFUS1-GFP. Transformation into E. coli. (Restriction digest, Gel electrophoresis, (Ligation, Transformation)
Overnights for Reporter Plasmids Assay: (Transformation)
- all cultures in SDmin medium
- Controls : 7283x, 7284a, 10150 a/x
- Samples in 7283a : pADH1, asCYC1, pSTE2, pSTE5, pTv3
- Samples in 7284x : pADH1, asCYC1, pSTE2, pSTE5, pTv3
- Samples in 10150a/x : pADH1, asCYC1, pSTE2, pSTE5, pTv3
PCR of reporter plasmids for biobricks. Restriction of reporter plasmids for biobricks. (PCR, Restriction digest)
No colonies on the plate with ligation of pRSII413+pFUS1-GFP.
pFUS1 assay
PCR of STE12. Restriction of STE12, pRSII406_MATa, pRSII406_MATx1x2. Ligation of STE12 into pRSII406_MATa and pRSII406_MATx1x2. Transformation into E. coli. (PCR, Restriction digest, Ligation, Transformation)
Colonies on both plates.
Summary: We prepared our reporter plasmids for biobricks assembly. Transformation of all reporter plasmids in yeast was successful. Assay was performed. We repeated ligation of STE12 with pRSII406_MATa and pRSII406_MATx1x2. Results will be next week.
Module 2
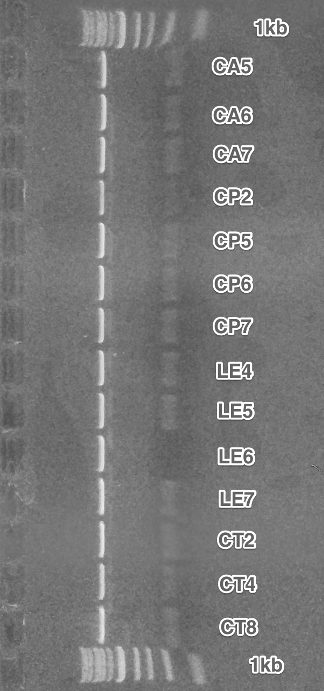
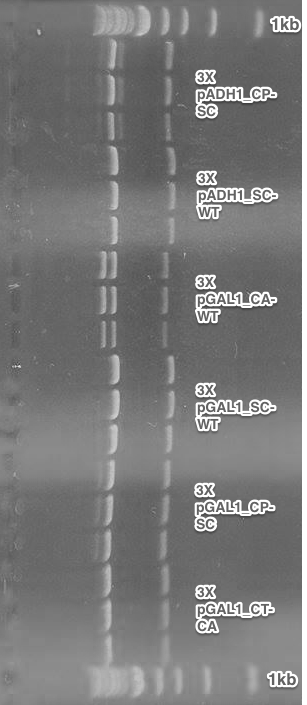
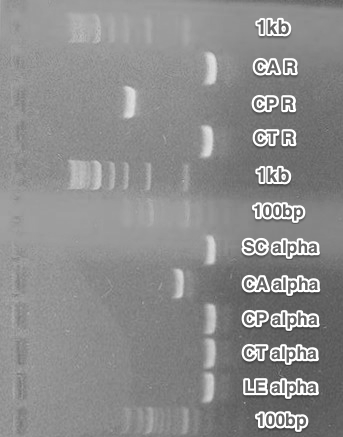
BioBricks preparation. All parts for the submission were amplified using PCR, that added the BioBrick restriction sites. After the restriction digest, these fragments were cloned into the pSB1C3 vectors.
The final parts were verified using Colony PCR.
We were successful with the CA receptor, CP-CA receptor and CA,CT,LE pheromones. SC and CP pheromones and CT receptor were not successfully verified.
Summary: We obtained 5 from 8 planned BioBricks.
Module 3
PCR amplification of the Aga1 gene from the genome and purification of the product (Genome extraction).
Restriction digest of yeast display plasmids(pCT302 Anti-cMyc/Anti-EpCAM/Anti-HuA/EpCAM/Streptavidin) and PCR product encoding Aga1 protein (Restriction digest).
Ligation of expression cassette and pRSII 413/415 plasmids (Ligation).
Transformation of DH5alpha bacteria cells, patches, minipreps and restriction digest testing (Transformation, Restriction digest).
Summary: We were not able to obtain correct expression vectors. However, we were successful at amplification of the Aga1 gene from genome and we also purified constructs encoding Aga2 and Anti-cMyc/Anti-EpCAM/Anti-HuA/EpCAM/Streptavidin fusion genes. Since our expression vector was not ready for cloning, we could not proceed.
Week 18 (31-06)
Module 1
Minipreps of pRSII406_MATa-STE12 ligation,pRSII406_MATx1x2-STE12 ligation. Verification restrictions and gel - unsuccessful. (Miniprep,Restriction digest, Gel electrophoresis)
Restriction of pRSII406_MATa (SnabI) and pRSII406_MATx1x2 (PstI) for integration. (Restriction digest)
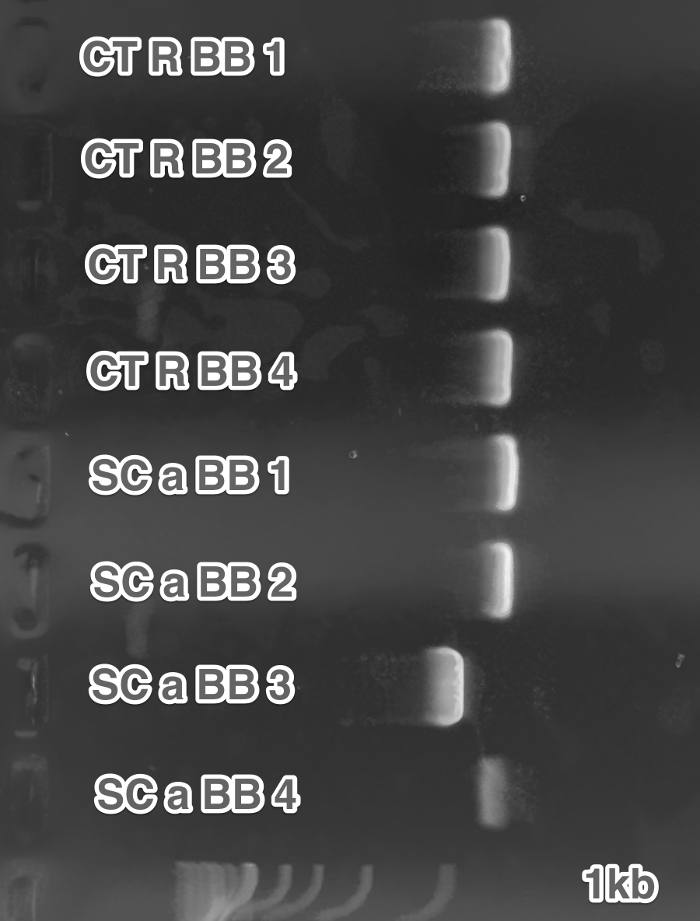
Overnight of 6193a, 6194x in YPD.(Transformation) Integration of pRSII406_MATa and pRSII406_MATx1x2.(Transformation)
Restriction of reporter plasmids. Ligation of reporters into 415(LEU), STE12 into 406. Transformstion into E. coli. (Restriction digest, Ligation, Transformation)
Plate results:
- colonies of STE12 in pRSII406
- no colonies of reporters in pRSII415
Overnight ligation of reporter plasmids into pRSII415. Transformation into E. coli. (Ligation, Transformation)
Minipreps of STE12. Verification restriction and gel of STE12 in pRSII406 - unsuccessful. (Miniprep, Restriction digest, Gel electrophoresis)
Plate results: reporters in pRSII415 - no colonies.
Restriction of reporters and pRSII415. Ligation of reporters into pRSII415. Transformation into E. coli. No colonies. (Restriction digest, Ligation, Transformation)
PCR of hybrid, as a template was the plasmid pH30 with primers whose added the terminator and without terminator. (PCR,Gel electrophoresis)
Using the same PCR conditions we did the bigger reaction for obtain more DNA. In the same time we do the band stab PCR for both bands. (PCR,Gel electrophoresis)
We extract all bands from gel. We cloned (restriction, ligation, transformation) the purified PCR hybrid constructs into pRSII413. So it was three ligations. We obtained one colony from the 2 kb band, do mini-prep and verification.(Restriction digest, Ligation, Transformation,Miniprep, Restriction digest, Gel electrophoresis)
Summary: We obtained plasmid with 2,1 kb long hybrid on pRSII413 with his auxotrophy. Ligation of STE12 in pRSII406 was unsuccessful. We were not albe to change auxotrophy of our reporter plasmids.
Module 2
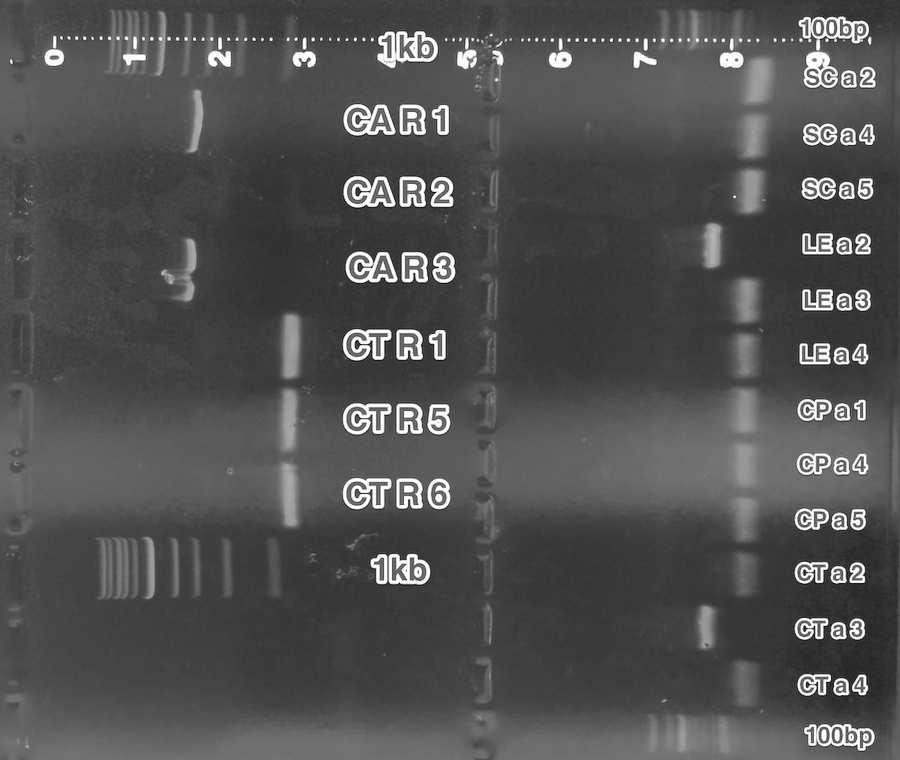
Remaining BioBricks were cloned again into pSB1C3 vectors.
We have verified SC pheromone but not the CT receptor.
Summary: We have created our sixth BioBrick.
Module 3
We transformed EBY100 yeast cells with Anti-cMyc, Anti-EpCAM, Anti-HuA and EpCAM inducible yeast display plasmids (Transformation).
Restriction of expression cassette and pRSII 413 and pRSII 415 vectors, subsequent gel purification, ligation and transformation of DH5alpha bacterial cells (Restriction digest, Ligation, Transformation).
Summary: Transformation of EBY100 yeast cells was not successful, no colonies were visible on the solid growing medium. The last attempt of universal expression plasmid (expression cassette on pRSII 413 and pRSII 415 plasmids) construction was not successfull.
Module 5
Signal transduction experiments in microfluidics: MATx + MATa pFUS1-GFP - done by Martin Cienciala.
September
Week 19 (07-13)
Module 1
We recieved new ligase and new ligase buffer.
Yeast transformation of reporter plasmids with GAL auxotrophy. (Transformation)
Restriction of reporter plasmids. Ligation of reporters into 415(LEU), MATa into 405, MATx into 405. Transformation of reporters in 415, MATa in 405, MATx in 405. (Restriction digest, LigationTransformation)
Plates results: colonies of MATa in 405, MATx in 405, pFUS1 in 415(LEU).
Colony PCS of MATa in 405, MATx in 405, pFUS1 in 415(LEU). Gel verification - all bands of correct size. (Colony PCR, Gel electrophoresis)
Overnight ligation of reporters into 415(LEU), MATa into 405, MATx into 405. (Ligation)
Transformation of overnight ligation of reporters in 415, MATa in 405, MATx in 405. Replating of ligation of reporters into 415(LEU). (Transformation)
Plates results: colonies on all plates.
Colony PCR of pSTE2-415, pSTE5-415, asCYC1-415. Gel verification - all bands of correct size.(Colony PCR, Gel electrophoresis)
Minipreps of pRSII415-MATa, pRSII415-MATx, pFUS1-415. Restriction. Gel verification - all bands of correct size. (Colony PCR, Gel electrophoresis)
Restriction of MATa (LEU) and MATx (LEU) - SnabI. Transformed MATa LEU and MATx LEU. (Restriction digest, Transformation)
Overnight ligation of biobricks - asCYC1, pTv3, pADH1, pSTE2. (Ligation)
Minipreps of pSTE2-415, pSTE5-415, asCYC1-415. Gel verification - all bands of correct size. (Miniprep, Restriction digest, Gel electrophoresis)
Transformation of biobricks. (Transformation)
Colony PCR verification of biobricks. Gel verification - most of the bands are correct. Patches of biobricks. (Colony PCR, Gel electrophoresis)
Mating of MATa(LEU) and MATx(LEU) yeasts. (Transformation)
Transformation of the plasmid with 2,1 kb long hybrid on pRSII413 into yeast strain 6194 MATalpha. (Transformation)
Summary: We successfuly changed auxotrophy of our synthetic MATa and MATx constructs. We were also able to clone our reporters into pRSII415 backbone. MATa and MATx were integrated into yeasts. Reporter plasmids were transformed into yeasts. We were not successful in yeast transformation of plasmid with hybrid.
We successfuly constructed 3 BioBricks and confirmed them with colony PCR. Gel verification will be done next week.
Module 2
Orthogonality assays using fluorescene as the output. Detailed description can be found on our project page.
Summary: We have verified the orthogonality of our receptors and signals. This is the major contribution from our module.
Module 3
PCR amplification of Anti-cMyc, Anti-EpCAM and EpCAM BioBricks from yeast display plasmids (PCR).
Purification of the PCR products, ligation and transformation to DH5alpha bacterial cells (Restriction digest, Ligation, Transformation).
Second attempt of yeast transformation with Anti-cMyc, Anti-EpCAM, Anti-HuA and EpCAM inducible yeast display plasmids (Transformation).
Immunostaining of yeast transformed with individual yeast display plasmids (Immunostaining).
Summary: We successfully transformed EBY100 yeast cells with four different types of yeast display plasmid. We also constructed the final BioBrick plasmids. Microscopy confirmed successful display of individual proteins.
Module 5
Bonding of PDMS to glass (3 microfluidic chips encapsulated on a glass slide) - done by Vaclav Pelisek.
Signal transduction experiments in microfluidics: MATx + MATa pFUS1-GFP - done by Martin Cienciala.
Week 20 (14-20)
Module 1
Minipreps of 3 biobricks. Restriction. Gel verification - successful. (Miniprep, Restriction digest, Gel electrophoresis)
Mating plan:
Assays - synsthetic MATa and MATx loci, reporter plasmids. (Transformation)
Module 3
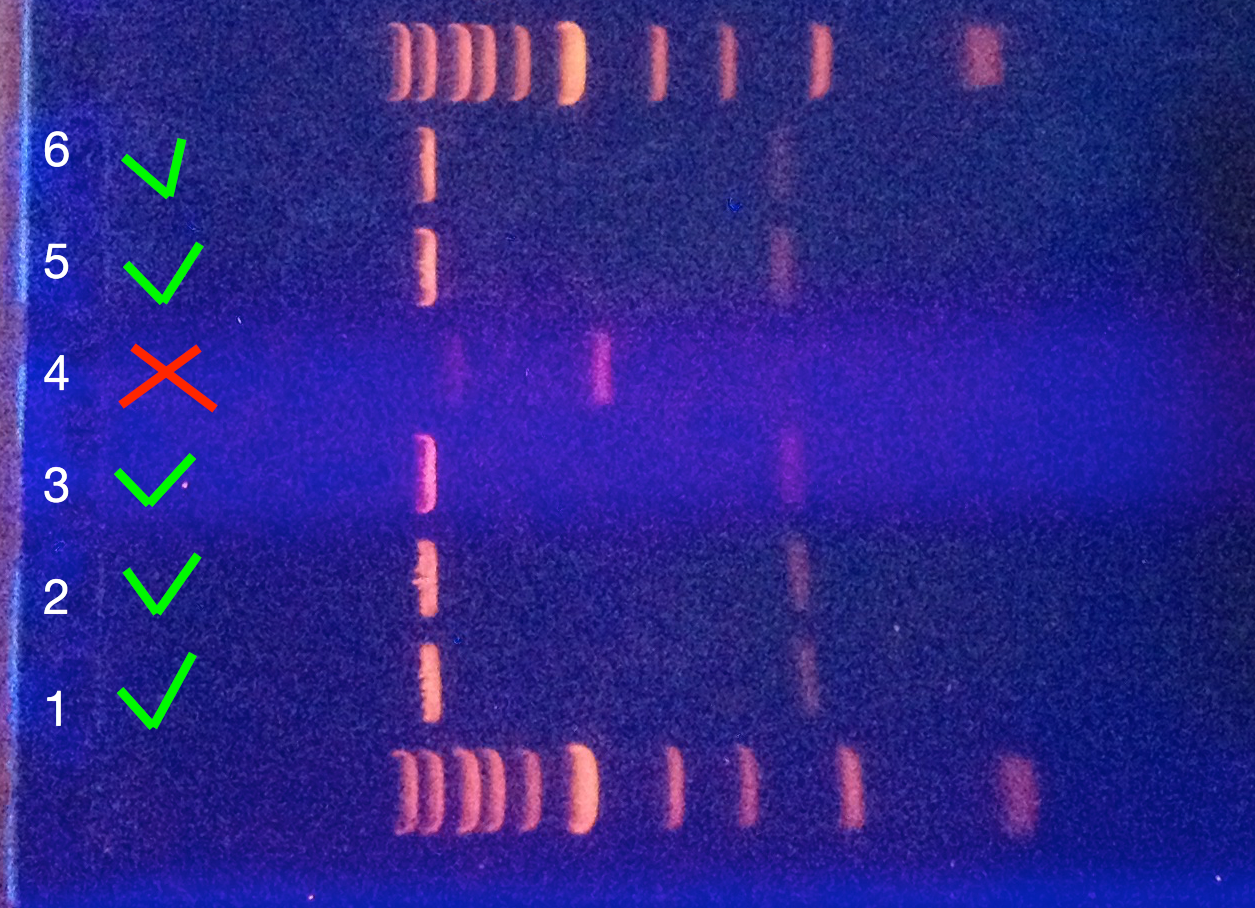
The strength of the Anti-EpCAM scFv to EpCAM bond formation was examined. Yeast cells (EBY100) transformed with yeast display plasmid containing EpCAM and Anti-EpCAM scFv inserts were grown in induction medium for 48 hours (20°C). After induction, cells were thoroughly vortexed to get rid of any pre-formed clumps. Samples containing only the negative control (displaying only Anti-EpCAM scFv) and samples containing both of the yeast types (yeast cells displaying anti-EpCAM scFv and yeast cells displaying EpCAM), were incubated for 5 minutes in 30°C. Solution of the medium and yeast cell aggregates was then qualitatively transferred onto a microscope slide, covered with a coverslip and examined.
We also ran an assay with yeast cells containing plasmids for copper inducible production of mating factor/display of Anti-EpCAM scFv on the surface of the yeast cell and cells containing plasmids for pheromone inducible production of GFP/display of EpCAM. We mixed these cells directly on the microreader (1:1) and induced them with the copper sulfate solution. We also had negative controls, which were cells transformed with the same plasmids except for the yeast display plasmids, therefore they could not bind each other.
Summary: We successfully observed clump formations in the examined samples. Fluorescence in the mixed culture was higher than in the cultures containing only one type of yeast.
Module 5
Bonding of PDMS to glass (6 microfluidic chips encapsulated on two glass slide) - done by Vaclav Pelisek.
Signal transduction experiments in microfluidics: synthetic MATa producing alpha-pheromone + MATa pFUS1-GFP - done by Martin Cienciala.
Yeast-induced blood agglutination on-chip - done by Martin Cienciala.
Note:
All pRSII plasmids were a gift from Steven Haase and were obtained from [http://www.addgene.org Addgene].
See Attributions to know who work on a specific parts of the project.