Difference between revisions of "Team:Brasil-USP/Project/Results"
Laisribovski (Talk | contribs) |
Laiscbrazaca (Talk | contribs) |
||
| (26 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{:Team:Brasil-USP/Templates/HeadContent|C1= | + | {{:Team:Brasil-USP/Templates/HeadContent|C1=Promoter and Exportantion tests|C2=Results}} |
<html> | <html> | ||
| Line 15: | Line 15: | ||
<li><a href="#promotertest">Promoter Test</a></li> | <li><a href="#promotertest">Promoter Test</a></li> | ||
<ul class="sub"> | <ul class="sub"> | ||
| − | <li><a href="#dh5alphabad"> | + | <li><a href="#dh5alphabad">DH5α</a></li> |
<li><a href="#BL21de3supercool">BL21 (DE3)</a></li> | <li><a href="#BL21de3supercool">BL21 (DE3)</a></li> | ||
<li><a href="#discussionpromotertest">Discussion</a></li> | <li><a href="#discussionpromotertest">Discussion</a></li> | ||
| Line 25: | Line 25: | ||
<li><a href="#discussionexportation">Discussion</a></li> | <li><a href="#discussionexportation">Discussion</a></li> | ||
</ul> | </ul> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br></ul> | <br></ul> | ||
| Line 46: | Line 39: | ||
<h1>Promoter Test</h1> | <h1>Promoter Test</h1> | ||
<p> | <p> | ||
| − | The Gram negative bacterium <i>Escherichia coli</i> is the most used chassis in Synthetic Biology. The bacterial system is often chosen due to its low cost, easy maintenance, high productivity and low time consumption. Moreover, there are several large scale system established for <i>E. coli</i> use in industry and, since our project aim for industrial applications, we have chosen the E. coli system allied with the previously named secretion routes that might help increase enzymes' stability. However, the choice of the promoter system is not as simple as the chassis'. That being said, we designed circuits to test the viability of three different inducible promoters: Plac, Para and Prha (figure | + | The Gram negative bacterium <i>Escherichia coli</i> is the most used chassis in Synthetic Biology. The bacterial system is often chosen due to its low cost, easy maintenance, high productivity and low time consumption. Moreover, there are several large scale system established for <i>E. coli</i> use in industry and, since our project aim for industrial applications, we have chosen the E. coli system allied with the previously named secretion routes that might help increase enzymes' stability. However, the choice of the promoter system is not as simple as the chassis'. That being said, we designed circuits to test the viability of three different inducible promoters: Plac, Para and Prha (figure 1). <br/> |
| + | |||
| + | <div class="fig" style="width: 1000px;"> | ||
| + | <img style="width:800px; display: block; margin: 20px auto 20px auto;" src="https://static.igem.org/mediawiki/2015/d/d0/Team-Brasil-USP_PromoterTestCircuits.png" /> | ||
| + | |||
| + | <p><center><font size=3>Figure 1 - Promoter test circuits (more information about the design of the circuits <a href="https://2015.igem.org/Team:Brasil-USP/Project/Design">here</a>). </center></font></p> | ||
| + | </div> | ||
| + | |||
</p> | </p> | ||
| − | + | ||
| − | + | ||
| − | + | ||
<a class="manchor" id="dh5alphabad"></a> | <a class="manchor" id="dh5alphabad"></a> | ||
| − | + | <h2>DH5α</h2> | |
| − | + | <p> We first started with DH5α. For protocols, we refer to our <a href="https://2015.igem.org/Team:Brasil-USP/Notebook/protocols">Protocols section</a>. We evaluated the fluorescence per cell by measuring Optical Density (OD) of our colonies and their fluorescences. We show in figures 2 and 3 our results.</p> | |
| − | + | ||
| − | + | ||
| − | + | <div class="fig" style="width: 600px;"> | |
| − | + | <img style="width:380px; display: block; margin: 20px auto 20px auto;" src="/wiki/images/8/89/Team-Brasil-USP_PTest_29_OD.png" /> | |
| − | + | ||
| − | + | <p><center><font size=3>Figure 2 - Optical Density (OD) measurements over time, showing that all colonies were in their log phase.</center></font></p> | |
| − | + | </div> | |
| + | |||
| + | <div class="fig" style="width: 750px;"> | ||
| + | <img style="width:750px; display: block; margin: 20px auto 20px auto;" src="https://static.igem.org/mediawiki/2015/d/df/Team-Brasil-USP_PTest_29_Results.png" /> | ||
| + | |||
| + | <p><font size=3><center>Figure 3 - Fluorescence per cell for each promoter, normalized by the fluorescence with no induction. In the x-axis, the higher the concentration label, the higher the concentration, with zero means no induction at all. Only Rhamnose showed some actual induction. All errors may be considered about 0.05 a.u.</center></font></p> | ||
| + | </div> | ||
| + | |||
| + | <p> Except for Rhamose, fluorescence was very weak and basically at the level of our negative control (E. Coli without plasmid), confirmed by a Mann-Whitney U test. Rhamnose showed some level of activity which would be considered weak, compared to, for instance, our <a href="/Team:Brasil-USP/interlabstudy">Interlab Results</a>. Finally, no considerable changes were detected in the RFP measures. Since there results are not good indicatives, we performed this experiment three different times (to reduce possible experimental artifacts with assembling or other steps in our protocol). Yet we have always observed the same behavior consistently.</p> | ||
<a class="manchor" id="bl21de3supercool"></a> | <a class="manchor" id="bl21de3supercool"></a> | ||
| − | + | <h2>BL21 (DE3)</h2> | |
| − | + | <p> We performed the same tests in BL21 strain. For this test, Arabinose was not used as we couldn't confirm its transformation in our gels. For protocols, we refer to our <a href="https://2015.igem.org/Team:Brasil-USP/Notebook/protocols">Protocols section</a>. Again, our colonies were all in log phase (omitted) when we measured their fluorescence over time. Our results are in figure 4 and 5.</p> | |
| − | + | ||
| − | + | ||
| + | <div class="fig" style="width: 750px;"> | ||
| + | <img style="width:750px; display: block; margin: 20px auto 20px auto;" src="/wiki/images/0/09/Team-Brasil-USP_PTest_Lac_GFP_RFP.png" /> | ||
| + | |||
| + | <p><center><font size=3>Figure 4 - Fluorescence per cell for pLac, normalized by the fluorescence with no induction. We see that with IPTG, GFP activity incresases and RFP activity decreases, as it should happen.</font></center></p> | ||
| + | </div> | ||
| − | < | + | <div class="fig" style="width: 750px;"> |
| − | + | <img style="width:750px; display: block; margin: 20px auto 20px auto;" src="/wiki/images/5/53/Team-Brasil-USP_PTest_Rham_GFP_RFP.png" /> | |
| − | + | ||
| − | + | <p><center><fon size=3>Figure 5 - Fluorescence per cell for each promoter, normalized by the fluorescence with no induction. Not only the induced activity is lower than the activity without induction - for the first 3 concentrations of Rhamnose -, but also the RFP is also growing.</center></font></p> | |
| − | + | ||
| − | + | ||
| + | <p> | ||
| + | <a href="http://parts.igem.org/Part:BBa_K1819008">BBa_K1819008</a> was submitted to the Registry of Standard Biological Parts as a Brasil-USP team new part. | ||
| + | </p> | ||
| + | </div> | ||
| + | <a class="manchor" id="discussionexportationtest"></a> | ||
| + | <h2>Discussion</h2> | ||
| + | <p> We successfully expressed and induced promoters pRha in DH5α strain and pLac in BL21 strain. Mann-Whitney U tests confirmed induction, i.e., induced fluorescence per cell was (statistically) significantly higher than our negative controls. Yet, the other promoters weren't successfully expressed or induced, , this fact can be explained by a mutation in the sequence, since we weren’t able to perform the sequencing of our circuits. Another possibility for our negative results is the wrong ligations between the parts, that again we just confirmed by agarose gel instead of sequencing. Since we had no issues at all with any of our constitutive promoters (see for instance our <a href="/Team:Brasil-USP/interlabstudy">Interlab Study</a>, <a href="/Team:Brasil-USP/Parts/KI">Pveg characterization in E. Coli</a>, <a href="/Team:Brasil-USP/Parts/Teamparts">Linker-GFP part</a>), our primary concern is that our protocols should be modified and optimized for induced promoters.</p> | ||
| Line 87: | Line 99: | ||
<h1>Exportation Test</h1> | <h1>Exportation Test</h1> | ||
<p> | <p> | ||
| − | + | TAs mentioned earlier, our enzymes have to be secreted so they can be in contact with the substrate, moreover it’s been seen that the heterologous expression of RoxA wasn’t successful due to inclusion body formation. The exportation of RoxA ensures its passage through the bacterial periplasm which increases the chances of a correct folding of the protein. Therefor we planned circuits with two exportation systems, the Twin-arginine translocation (TAT) sequence and the fusion of the enzymes with the bacterial outer membrane, OmpA, to check which of these exportation systems were better for each of the enzymes. The designed circuits are shown in the figure below (Figure 6). <br/> | |
| − | + | ||
| + | <div class="fig" style="width: 800px;"> | ||
| + | <img style="width:500px; display: block; margin: 20px auto 20px auto;" src="https://static.igem.org/mediawiki/2015/7/7e/Team-Brasil-USP_ExportationTestCircuits.png" /> | ||
| + | |||
| + | <p><center><font size=3>Figure 6 - Exportation test circuits (more information about the design of the circuits <a href="https://2015.igem.org/Team:Brasil-USP/Project/Design">here</a>). </center></font></p> | ||
| + | </div> | ||
| + | </p> | ||
<a class="manchor" id="tatsignal"></a> | <a class="manchor" id="tatsignal"></a> | ||
<h2>TAT signal</h2> | <h2>TAT signal</h2> | ||
<p> | <p> | ||
| − | The Twin-arginine translocation (TAT) system is classified as part of the type II bacterial secretion mechanism, which is composed of two steps. The first one aims to get the protein through the bacterial cytoplasmic membrane and, for that, it can use three different pathways (the SecB-dependent pathway, the signal recognition particle (SRP), and the twin-arginine translocation (TAT)). The second step is the protein translocation through the outer membrane, involving specific protein machinery known as the secreton.<sup>5</sup> <br> | + | The Twin-arginine translocation (TAT) system is classified as part of the type II bacterial secretion mechanism, which is composed of two steps. The first one aims to get the protein through the bacterial cytoplasmic membrane and, for that, it can use three different pathways (the SecB-dependent pathway, the signal recognition particle (SRP), and the twin-arginine translocation (TAT)). The second step is the protein translocation through the outer membrane, involving specific protein machinery known as the secreton.<sup>5</sup> <br> |
| − | The TAT pathway, unlike the SecB or the SRP pathways, is capable of transporting folded proteins across the inner membrane independently of ATP using the transmembrane PMF (proton-motive force).<sup>5</sup> This translocation system is composed mainly of five proteins (TatA, TatB, TatC, TatD and TatE) but their specific function have not been firmly established yet. However, it is known that TatA is the most expressed gene of this pathway and it has 60% of homology with TatE, which can partially substitute TatA; TaTB can form a complex with TatC to prevent its degradation; there is evidence that TatC is the signal peptide binding protein and, finally, TatD produces no effect on protein translocation but has DNase activity.5 The secretion is composed by 12-16 different proteins, but none of these proteins have their specific roles elucidated.<sup>5</sup> | + | The TAT pathway, unlike the SecB or the SRP pathways, is capable of transporting folded proteins across the inner membrane independently of ATP using the transmembrane PMF (proton-motive force).<sup>5</sup> This translocation system is composed mainly of five proteins (TatA, TatB, TatC, TatD and TatE) but their specific function have not been firmly established yet. However, it is known that TatA is the most expressed gene of this pathway and it has 60% of homology with TatE, which can partially substitute TatA; TaTB can form a complex with TatC to prevent its degradation; there is evidence that TatC is the signal peptide binding protein and, finally, TatD produces no effect on protein translocation but has DNase activity.<sup>5</sup> The secretion is composed by 12-16 different proteins, but none of these proteins have their specific roles elucidated.<sup>5</sup> |
</p> | </p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<a class="manchor" id="ompa"></a> | <a class="manchor" id="ompa"></a> | ||
<h2>OmpA</h2> | <h2>OmpA</h2> | ||
<p> | <p> | ||
| − | The ompA mechanism is inserted in the type I secretion system which, in Gram negative bacteria, promotes the secretion of various proteins of different sizes and functions in one single step, without the necessity of a periplasmic intermediate. The secretion is driven by three proteins of the cell | + | The ompA mechanism is inserted in the type I secretion system which, in Gram negative bacteria, promotes the secretion of various proteins of different sizes and functions in one single step, without the necessity of a periplasmic intermediate. The secretion is driven by three proteins of the cell envelope<sup>6</sup>. The first one is located in the outer membrane, and is called OMP (Outer Membrane Protein); the other two are cytoplasmic membrane proteins, called ABC (ATP-Binding Cassette) and MFP (Membrane Fusion Protein). The protein being transported has a specific C-terminal signal sequence that usually has few to 50 glycine rich-repeats, that might be necessary for the protein function and, therefore, is not cleaved during the secretion6. This signal sequence recognizes the ABC protein and triggers the assembly of the whole complex for the translocation process. |
</p> | </p> | ||
| Line 114: | Line 126: | ||
<h2>Discussion</h2> | <h2>Discussion</h2> | ||
<p> | <p> | ||
| − | + | We intended to evaluate the exportations systems through GFP fluorescence, since this phenomenon depends on the correct folding of the protein, which in this case is linked with the folding of Lcp and RoxA. Unfortunately we weren’t able to perform these tests, first due to our problems with promoter test and second because we are still in the circuit construction phase. | |
</p> | </p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<a class="manchor" id=""></a> | <a class="manchor" id=""></a> | ||
| Line 163: | Line 138: | ||
4. Bode, H. B., Kerkhoff, K. & Jendrossek, D. Bacterial degradation of natural and synthetic rubber. Biomacromolecules 2, 295-303, doi:10.1021/bm005638h (2001).<br> | 4. Bode, H. B., Kerkhoff, K. & Jendrossek, D. Bacterial degradation of natural and synthetic rubber. Biomacromolecules 2, 295-303, doi:10.1021/bm005638h (2001).<br> | ||
5. Mergulhão, F. J. M.; Summers, D. K.; Monteiro, G. A. Recombinant protein secretion in Escherichia coli. Biotechnology Advances. 23, 177–202 (2005).<br> | 5. Mergulhão, F. J. M.; Summers, D. K.; Monteiro, G. A. Recombinant protein secretion in Escherichia coli. Biotechnology Advances. 23, 177–202 (2005).<br> | ||
| − | 6. Delepelaire, P. Type I secretion in | + | 6. Delepelaire, P. Type I secretion in g |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 13:48, 15 November 2015
Promoter and Exportantion tests
Results

Table of contents
Natural and synthetic rubber degradation has been reported for some microorganisms such as Streptomyces sp. strain K30, Gordonia polyisoprenivorans, Nocardia sp. strain 835A and Xanthomonas sp. strain 35Y.1 However, most of these organisms show a slow growth when using rubber as a sole carbon source.2-4
To improve microbial rubber degradation efficiency, our project aims to create a gene circuit which will, among other things, be responsible for coding and expressing two enzymes: RoxA (Rubber oxygenase A) and Lcp (Latex clearing protein). These enzymes are fundamental for rubber degradation and have to be secreted or exposed in the exterior of the cell to be in direct contact with their substrate, the poly(cis-1,4 isoprene) - the main component of rubber. The proposed circuit provides the secretion of the proteins by the fusion with a specific signal sequence, the Twin-arginine translocation (TAT) sequence, or its binding to the bacterial outer membrane by the fusion with a specific protein, OmpA fused with a linker (BBa_K1489002).
Promoter Test
The Gram negative bacterium Escherichia coli is the most used chassis in Synthetic Biology. The bacterial system is often chosen due to its low cost, easy maintenance, high productivity and low time consumption. Moreover, there are several large scale system established for E. coli use in industry and, since our project aim for industrial applications, we have chosen the E. coli system allied with the previously named secretion routes that might help increase enzymes' stability. However, the choice of the promoter system is not as simple as the chassis'. That being said, we designed circuits to test the viability of three different inducible promoters: Plac, Para and Prha (figure 1).

DH5α
We first started with DH5α. For protocols, we refer to our Protocols section. We evaluated the fluorescence per cell by measuring Optical Density (OD) of our colonies and their fluorescences. We show in figures 2 and 3 our results.
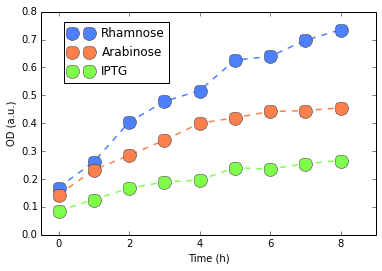

Except for Rhamose, fluorescence was very weak and basically at the level of our negative control (E. Coli without plasmid), confirmed by a Mann-Whitney U test. Rhamnose showed some level of activity which would be considered weak, compared to, for instance, our Interlab Results. Finally, no considerable changes were detected in the RFP measures. Since there results are not good indicatives, we performed this experiment three different times (to reduce possible experimental artifacts with assembling or other steps in our protocol). Yet we have always observed the same behavior consistently.
BL21 (DE3)
We performed the same tests in BL21 strain. For this test, Arabinose was not used as we couldn't confirm its transformation in our gels. For protocols, we refer to our Protocols section. Again, our colonies were all in log phase (omitted) when we measured their fluorescence over time. Our results are in figure 4 and 5.
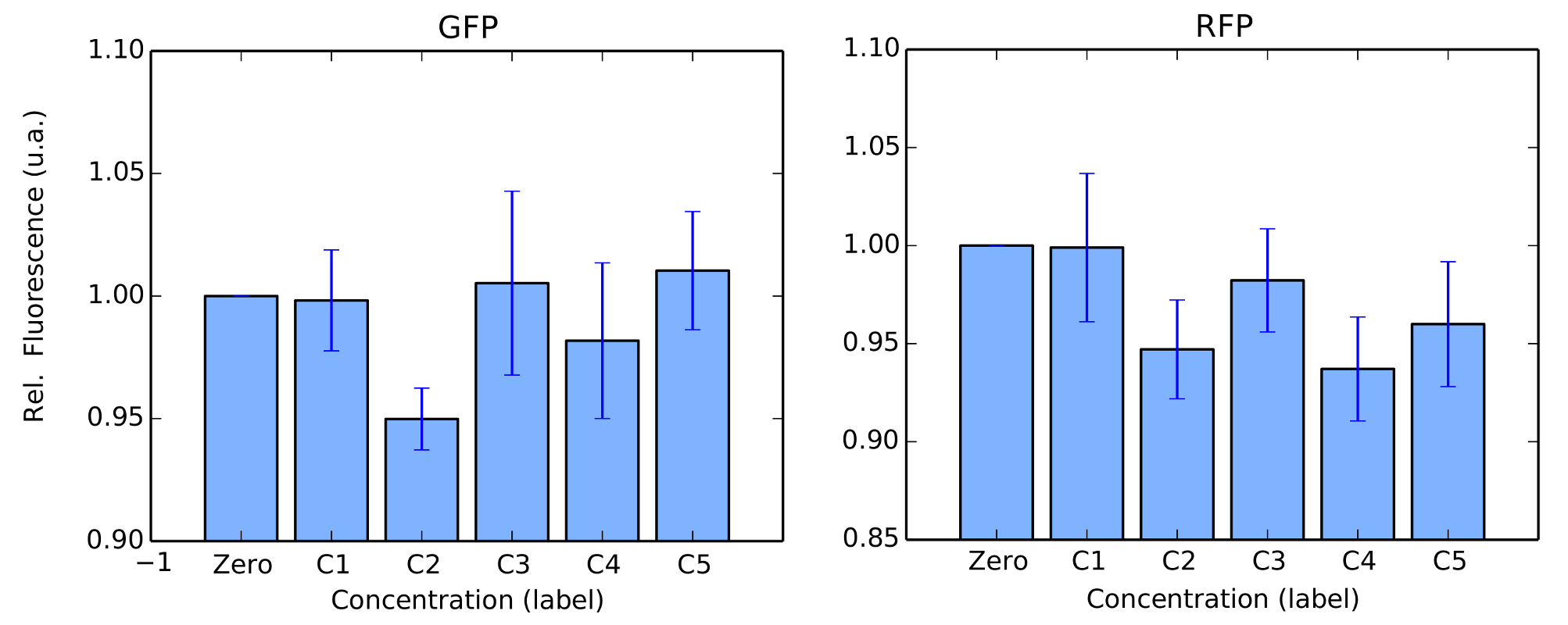
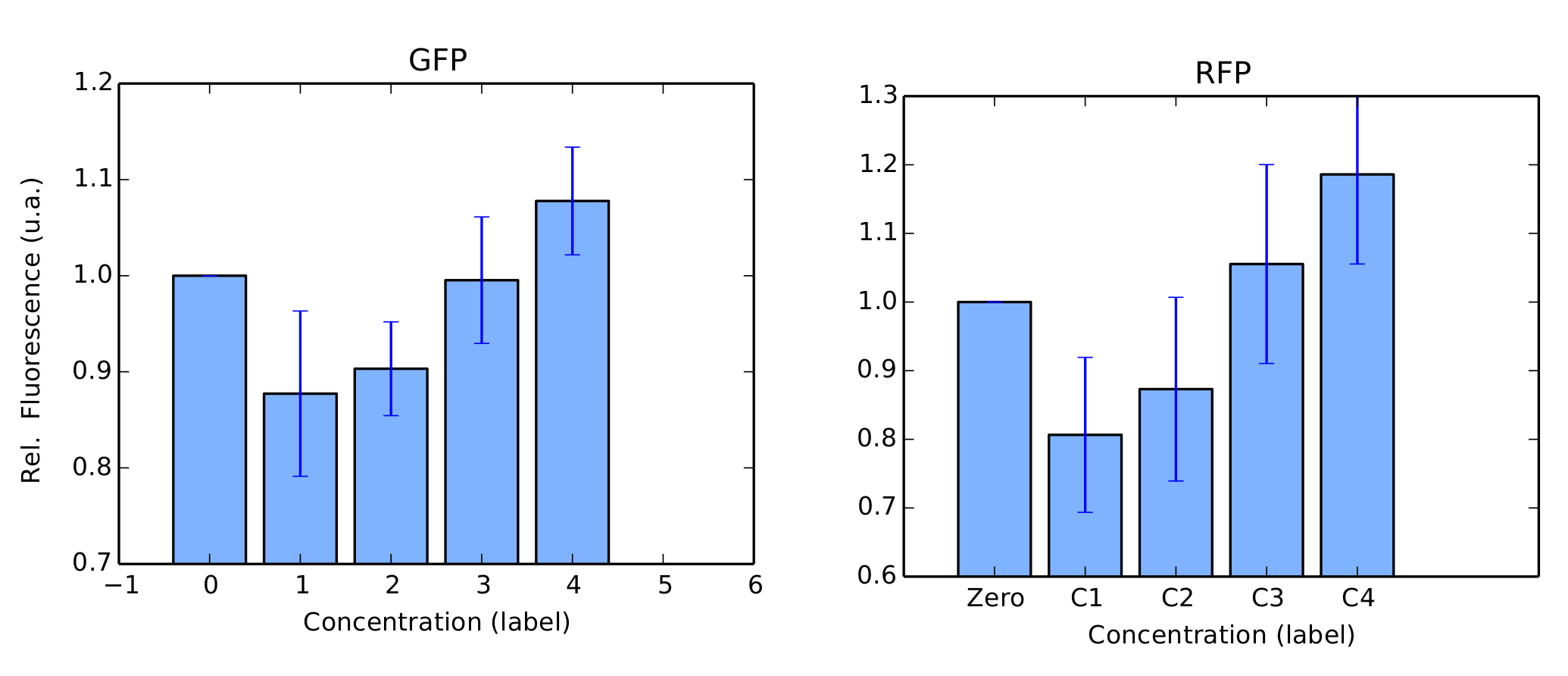
BBa_K1819008 was submitted to the Registry of Standard Biological Parts as a Brasil-USP team new part.
Discussion
We successfully expressed and induced promoters pRha in DH5α strain and pLac in BL21 strain. Mann-Whitney U tests confirmed induction, i.e., induced fluorescence per cell was (statistically) significantly higher than our negative controls. Yet, the other promoters weren't successfully expressed or induced, , this fact can be explained by a mutation in the sequence, since we weren’t able to perform the sequencing of our circuits. Another possibility for our negative results is the wrong ligations between the parts, that again we just confirmed by agarose gel instead of sequencing. Since we had no issues at all with any of our constitutive promoters (see for instance our Interlab Study, Pveg characterization in E. Coli, Linker-GFP part), our primary concern is that our protocols should be modified and optimized for induced promoters.
Exportation Test
TAs mentioned earlier, our enzymes have to be secreted so they can be in contact with the substrate, moreover it’s been seen that the heterologous expression of RoxA wasn’t successful due to inclusion body formation. The exportation of RoxA ensures its passage through the bacterial periplasm which increases the chances of a correct folding of the protein. Therefor we planned circuits with two exportation systems, the Twin-arginine translocation (TAT) sequence and the fusion of the enzymes with the bacterial outer membrane, OmpA, to check which of these exportation systems were better for each of the enzymes. The designed circuits are shown in the figure below (Figure 6).

TAT signal
The Twin-arginine translocation (TAT) system is classified as part of the type II bacterial secretion mechanism, which is composed of two steps. The first one aims to get the protein through the bacterial cytoplasmic membrane and, for that, it can use three different pathways (the SecB-dependent pathway, the signal recognition particle (SRP), and the twin-arginine translocation (TAT)). The second step is the protein translocation through the outer membrane, involving specific protein machinery known as the secreton.5
The TAT pathway, unlike the SecB or the SRP pathways, is capable of transporting folded proteins across the inner membrane independently of ATP using the transmembrane PMF (proton-motive force).5 This translocation system is composed mainly of five proteins (TatA, TatB, TatC, TatD and TatE) but their specific function have not been firmly established yet. However, it is known that TatA is the most expressed gene of this pathway and it has 60% of homology with TatE, which can partially substitute TatA; TaTB can form a complex with TatC to prevent its degradation; there is evidence that TatC is the signal peptide binding protein and, finally, TatD produces no effect on protein translocation but has DNase activity.5 The secretion is composed by 12-16 different proteins, but none of these proteins have their specific roles elucidated.5
OmpA
The ompA mechanism is inserted in the type I secretion system which, in Gram negative bacteria, promotes the secretion of various proteins of different sizes and functions in one single step, without the necessity of a periplasmic intermediate. The secretion is driven by three proteins of the cell envelope6. The first one is located in the outer membrane, and is called OMP (Outer Membrane Protein); the other two are cytoplasmic membrane proteins, called ABC (ATP-Binding Cassette) and MFP (Membrane Fusion Protein). The protein being transported has a specific C-terminal signal sequence that usually has few to 50 glycine rich-repeats, that might be necessary for the protein function and, therefore, is not cleaved during the secretion6. This signal sequence recognizes the ABC protein and triggers the assembly of the whole complex for the translocation process.
Discussion
We intended to evaluate the exportations systems through GFP fluorescence, since this phenomenon depends on the correct folding of the protein, which in this case is linked with the folding of Lcp and RoxA. Unfortunately we weren’t able to perform these tests, first due to our problems with promoter test and second because we are still in the circuit construction phase.
References
1. Rose, K. & Steinbuchel, A. Biodegradation of natural rubber and related compounds: Recent insights into a hardly understood catabolic capability of microorganisms. Appl. Environ. Microbiol. 71, 2803-2812, doi:10.1128/aem.71.6.2803-2812.2005 (2005).
2. Rose, K., Tenberge, K. B. & Steinbuchel, A. Identification and characterization of genes from Streptomyces sp strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules 6, 180-188, doi:10.1021/bm0496110 (2005).
3. Tsuchii, A. & Takeda, K. Rubber-degrading enzyme from a bacterial culture. Appl. Environ. Microbiol. 56, 269-274 (1990).
4. Bode, H. B., Kerkhoff, K. & Jendrossek, D. Bacterial degradation of natural and synthetic rubber. Biomacromolecules 2, 295-303, doi:10.1021/bm005638h (2001).
5. Mergulhão, F. J. M.; Summers, D. K.; Monteiro, G. A. Recombinant protein secretion in Escherichia coli. Biotechnology Advances. 23, 177–202 (2005).
6. Delepelaire, P. Type I secretion in g