Difference between revisions of "Team:UCLA/Notebook/Honeybee Silk/13 July 2015"
(→Visualization) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 298: | Line 298: | ||
|- | |- | ||
! Component | ! Component | ||
| − | ! Volume (out of | + | ! Volume (out of 25uL) |
|- | |- | ||
! APEX Taq Red Mastermix | ! APEX Taq Red Mastermix | ||
| Line 316: | Line 316: | ||
| 9uL | | 9uL | ||
|} | |} | ||
| + | |||
| + | |||
| + | ===Program=== | ||
| + | {| class="wikitable" style="text-align:center; width:600px; height:200px;" | ||
| + | |+ | ||
| + | |- | ||
| + | ! Step | ||
| + | ! Temperature | ||
| + | ! Time | ||
| + | |- | ||
| + | ! Initial Denaturation | ||
| + | | 95C | ||
| + | | 30s | ||
| + | |- | ||
| + | |||
| + | ! Cycles(x30) | ||
| + | | 95C | ||
| + | | 10s | ||
| + | |- | ||
| + | ! Annealing | ||
| + | | 67C | ||
| + | | 20s | ||
| + | |- | ||
| + | ! Extension | ||
| + | | 70C | ||
| + | | 30s | ||
| + | |- | ||
| + | !Final Extension | ||
| + | | 70C | ||
| + | | 2 min | ||
| + | |- | ||
| + | ! Hold | ||
| + | |14C | ||
| + | | Hold | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ='''Visualization'''= | ||
| + | |||
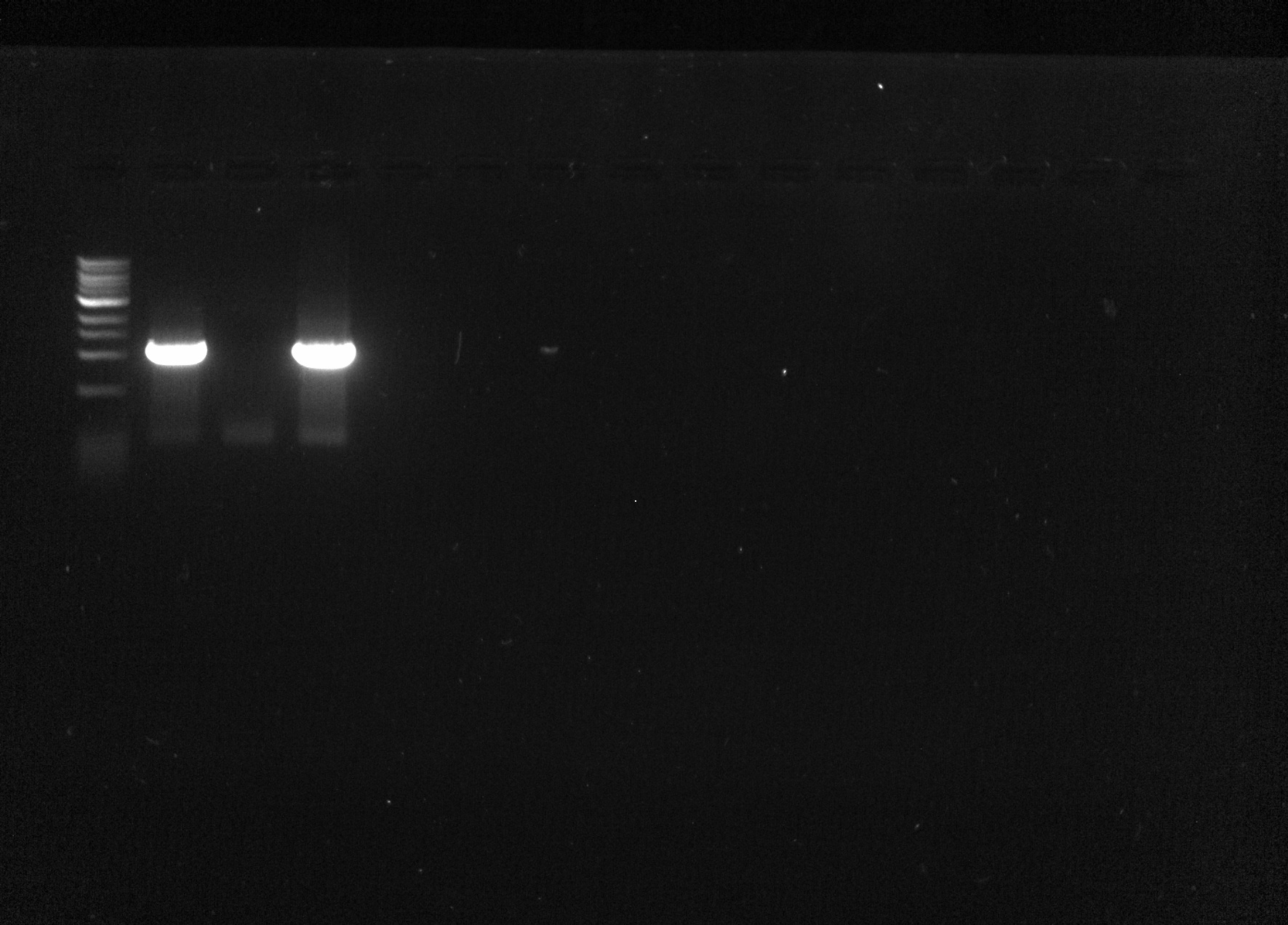
| + | I ran each of the sample on a 1% agarose gel against a 1kB ladder for 20 minutes. | ||
| + | Expected band sizes: ~1000bp | ||
| + | |||
| + | Samples 1 and 3 had bright bands at the appropriate base pair length. | ||
| + | |||
| + | [[File:Gel 2015-07-13.jpg|none|thumb|500px|Gel Image]] | ||
| + | |||
| + | ='''Inoculation'''= | ||
| + | |||
| + | The remaining 99uL of samples 1 and 3 were inoculated with 5mL LB broth and 5uL 1000X kanamycin, incubated at 37C. | ||
| + | |||
=Testing competency of BL21 cells from 7/12= | =Testing competency of BL21 cells from 7/12= | ||
*Used puc 19 vector at 50 picograms/ul to test transformation efficiency. | *Used puc 19 vector at 50 picograms/ul to test transformation efficiency. | ||
*Added 2 ul of puc 19 to 50 ul of competent BL21 cells | *Added 2 ul of puc 19 to 50 ul of competent BL21 cells | ||
*Used [https://www.neb.com/protocols/1/01/01/high-efficiency-transformation-protocol-c2987 this NEB protocol] to transform with our BL21 cells. | *Used [https://www.neb.com/protocols/1/01/01/high-efficiency-transformation-protocol-c2987 this NEB protocol] to transform with our BL21 cells. | ||
| + | *Plated 10% of the cells on a warm amp plate and put plate in at 4 pm 7/13 | ||
Latest revision as of 20:41, 15 July 2015
Contents
Colony PCR of 7/12 Transformation
- The transformation was successful for the 3:1 ligated products plates. I picked 3 colonies off the plate and suspended each in 100uL of ddH2O.
- Using APEX Taq Red Mastermix
| Component | Volume (out of 25uL) |
|---|---|
| APEX Taq Red Mastermix | 12.5uL |
| 10mM primer 10 | 1.25uL |
| 10mM primer 11 | 1.25uL |
| Transformed Cells | 1uL |
| Nuclease Free Water | 9uL |
Program
| Step | Temperature | Time |
|---|---|---|
| Initial Denaturation | 95C | 30s |
| Cycles(x30) | 95C | 10s |
| Annealing | 67C | 20s |
| Extension | 70C | 30s |
| Final Extension | 70C | 2 min |
| Hold | 14C | Hold |
Visualization
I ran each of the sample on a 1% agarose gel against a 1kB ladder for 20 minutes. Expected band sizes: ~1000bp
Samples 1 and 3 had bright bands at the appropriate base pair length.
Inoculation
The remaining 99uL of samples 1 and 3 were inoculated with 5mL LB broth and 5uL 1000X kanamycin, incubated at 37C.
Testing competency of BL21 cells from 7/12
- Used puc 19 vector at 50 picograms/ul to test transformation efficiency.
- Added 2 ul of puc 19 to 50 ul of competent BL21 cells
- Used this NEB protocol to transform with our BL21 cells.
- Plated 10% of the cells on a warm amp plate and put plate in at 4 pm 7/13