Difference between revisions of "Team:Czech Republic/Project/Orthogonal signals and receptors"
(→Receptors) |
(→Receptors) |
||
| Line 68: | Line 68: | ||
Receptors were constructed by a restriction/ligation using pRSII416 vector. Wild-type STE2 from ''Saccharomyces cerevisiae'' was obtained by a PCR from the genome (the sequence can be found as [http://www.yeastgenome.org/locus/S000001868/overview YFL026W]), pGAL1 promoter was obtained from the Registry (part [http://parts.igem.org/Part:BBa_J63006 BBa_J63006]). Other STE2 receptors were ordered as gBlocks. | Receptors were constructed by a restriction/ligation using pRSII416 vector. Wild-type STE2 from ''Saccharomyces cerevisiae'' was obtained by a PCR from the genome (the sequence can be found as [http://www.yeastgenome.org/locus/S000001868/overview YFL026W]), pGAL1 promoter was obtained from the Registry (part [http://parts.igem.org/Part:BBa_J63006 BBa_J63006]). Other STE2 receptors were ordered as gBlocks. | ||
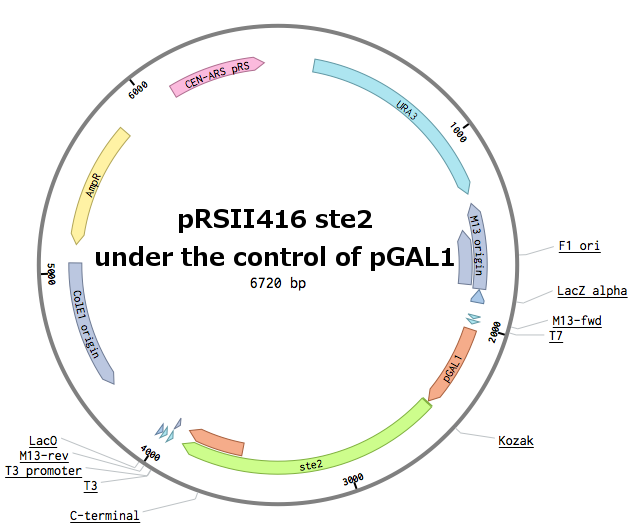
| − | First, the plasmid pGAL1_SC_WT was constructed | + | First, the plasmid pGAL1_SC_WT was constructed: |
[[File:Czech_Republic_Ste2FinalScheme.png|350px|Scheme of the final plasmid]] | [[File:Czech_Republic_Ste2FinalScheme.png|350px|Scheme of the final plasmid]] | ||
Revision as of 12:32, 14 September 2015
Synthetic signals and receptors
Contents
Abstract
In order to achieve more complex behaviour of our IODs, each IOD needs to be able to communicate with others. Since the pheromone response pathway was used for signal transduction, we prepared several synthetic signals and receptors that couple to this pathway and allow our IODs to communicate without a cross-talk.
Key Achievements
- Constructed a set of plasmids with different pheromones and their receptors.
- Verified the correct coupling of the receptors to the pheromone mating pathway.
- Verified the correct expression and secretion of the different pheromones.
Intro / Background
Test [Janiak2005]. [Lin2011]
Design
Concept
Receptors
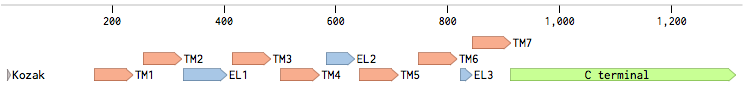
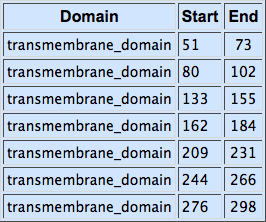
Alpha factor receptor (STE2) is produced and displayed by a cells. It is a 431aa long seven transmembrane-domain (TM in the figure) GPCR. Since the sequence is known, it can be easily obtained by a PCR from the genome of our Saccharomyces cerevisiae strain. For higher translation in yeast, we added consensus Kozak sequence before the start codon. Also, restriction sites were added to the ends of the fragment to allow easy future cloning.
Other STE2 receptors (STE2 receptors from different yeast strains) were obtained via gBlocks® order. In order to achieve effective coupling to the pheromone mating pathway in the Saccharomyces cerevisiae strain and proper expression, we performed several tasks.
First, after the initial protein [http://blast.ncbi.nlm.nih.gov/Blast.cgi BLAST] search, codon optimization was done using an online tool from IDT. During this proccess, all BioBrick restriction sites were removed from the DNA sequence.
Second, for each of the STE2 receptor, its C-terminal was replaced by the C-terminal from Candida albicans or Saccharomyces cerevisiae in order to improve coupling to the pheromone mating pathway [Lin2011]. The effect of each replacement on the protein folding was checked using the [http://elm.eu.org/ ELM resource]. Using this tool, we have verified that the changes in the C-terminal have no effect on the transmembrane domains.
Finally, the DNA sequences were optimized according to the recommendations from the IDT ordering form to speed up the synthesis process.
Signals
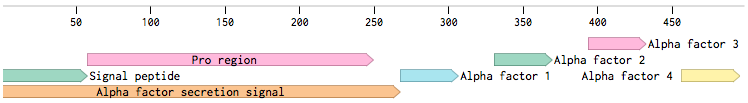
Alpha-factor mating pheromone is a short peptide (typically 13aa long) secreted by alpha cells. For our project, we needed to produce a variety of these pheromones and secrete them from the cells. Altough there are some secretion tags present in the registry (BBa_K416003, BBa_K792002), there are no parts encoding pheromones available. Also, since we found evidence that the complete wild-type signal tag is needed for the proper secretion of the pheromone [Caplan1991], we decided to obtain the whole sequence directly from the MF(ALPHA)1 locus.
MF(ALPHA)1 gene codes for four mature alpha-factors within a putative precursor of 165 amino acids. This sequence begins with a signal tag for secretion and a segment with three glycosylation sites (89aa). The second segment contains four mature alpha-factors, each preceded by spacer peptides, which contain proteolytic processing signals [Kurjan1982].
Since we wanted the pheromone peptides to be easily changeable by Site-directed mutagenesis (SDM), we decided to preserve the signal tag with only one copy of the pheromone:
This would create a unique site for the SDM. The final production level of the pheromone is supposed to be practically the same, since the used plasmid is present in the cell in more copies (2-5). Also, for higher expression in the cells, we added consensus Kozak sequence before the start codon. Restriciton sites were added to the ends of the sequence for easy future cloning.
DNA
Receptors
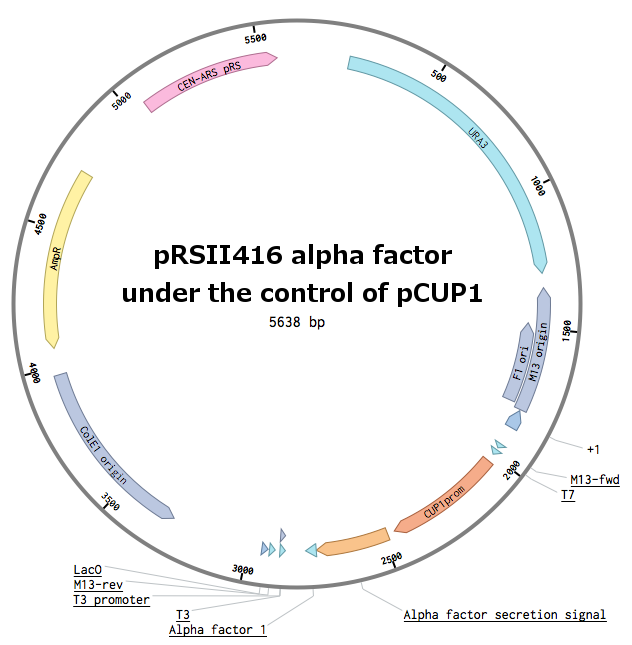
Receptors were constructed by a restriction/ligation using pRSII416 vector. Wild-type STE2 from Saccharomyces cerevisiae was obtained by a PCR from the genome (the sequence can be found as [http://www.yeastgenome.org/locus/S000001868/overview YFL026W]), pGAL1 promoter was obtained from the Registry (part [http://parts.igem.org/Part:BBa_J63006 BBa_J63006]). Other STE2 receptors were ordered as gBlocks.
First, the plasmid pGAL1_SC_WT was constructed:
Saccharomyces cerevisiae
ATGTCTGATGCGGCTCCTTCATTGAGCAATCTATTTTATGATCCAACGTATAATCCTGGTCAAAGCACCATTAACTACACTTCCATATATGGGAATGGATCTACCATCACTTTCGATGAGTTGCAAGGTTTAGTTAACAGTACTGTTACTCAGGCCATTATGTTTGGTGTCAGATGTGGTGCAGCTGCTTTGACTTTGATTGTCATGTGGATGACATCGAGAAGCAGAAAAACGCCGATTTTCATTATCAACCAAGTTTCATTGTTTTTAATCATTTTGCATTCTGCACTCTATTTTAAATATTTACTGTCTAATTACTCTTCAGTGACTTACGCTCTCACCGGATTTCCTCAGTTCATCAGTAGAGGTGACGTTCATGTTTATGGTGCTACAAATATAATTCAAGTCCTTCTTGTGGCTTCTATTGAGACTTCACTGGTGTTTCAGATAAAAGTTATTTTCACAGGCGACAACTTCAAAAGGATAGGTTTGATGCTGACGTCGATATCTTTCACTTTAGGGATTGCTACAGTTACCATGTATTTTGTAAGCGCTGTTAAAGGTATGATTGTGACTTATAATGATGTTAGTGCCACCCAAGATAAATACTTCAATGCATCCACAATTTTACTTGCATCCTCAATAAACTTTATGTCATTTGTCCTGGTAGTTAAATTGATTTTAGCTATTAGATCAAGAAGATTCCTTGGTCTCAAGCAGTTCGATAGTTTCCATATTTTACTCATAATGTCATGTCAATCTTTGTTGGTTCCATCGATAATATTCATCCTCGCATACAGTTTGAAACCAAACCAGGGAACAGATGTCTTGACTACTGTTGCAACATTACTTGCTGTATTGTCTTTACCATTATCATCAATGTGGGCCACGGCTGCTAATAATGCATCCAAAACAAACACAATTACTTCAGACTTTACAACATCCACAGATAGGTTTTATCCAGGCACGCTGTCTAGCTTTCAAACTGATAGTATCAACAACGATGCTAAAAGCAGTCTCAGAAGTAGATTATATGACCTATATCCTAGAAGGAAGGAAACAACATCGGATAAACATTCGGAAAGAACTTTTGTTTCTGAGACTGCAGATGATATAGAGAAAAATCAGTTTTATCAGTTGCCCACACCTACGAGTTCAAAAAATACTAGGATAGGACCGTTTGCTGATGCAAGTTACAAAGAGGGAGAAGTTGAACCCGTCGACATGTACACTCCCGATACGGCAGCTGATGAGGAAGCCAGAAAGTTCTGGACTGAAGATAATAATAATTTATGA
Primers
| Name | Sequence |
|---|---|
| ig2-ste2p-HindIII-F | tcaatgaagcttTACAAAATGTCTGATGCGGCTCCT |
| ig2-ste2p-BamHI-R | tcagatggatccTCATAAATTATTATTATCTTCAGTCCAGAACT |
| ig2-pGAL1-XhoI-F | tacagcctcgagCGGATTAGAAGCCGCCGAG |
| ig2-pGAL1-HindIII-R | tcacgtaagcttCTCCTTGACGTTAAAGTATAGAGGT |
gBlocks with Kozak, restriction sites and sites for M13 primers
Candida albicans with its (CA) C-terminal
gtaaaacgacggccagtctcgagaagcttTACAAAATGAACATAAATTCTACATTCATTCCAGACAAGCCAGGGGATATCATTATTAGTTATAGTATACCGGGATTGGACCAGCCCATTCAAATACCATTCCATAGTTTAGATTCCTTTCAAACAGACCAAGCTAAGATCGCTTTAGTTATGGGTATCACAATAGGATCTTGTTCTATGACACTTATCTTTCTAATCTCAATCATGTATAAAACGAATAAGTTGACAAACTTGAAGTTGAAACTTAAGTTAAAGTACATCTTGCAATGGATCAACCAAAAGATCTTCACGAAGAAGAGAAATGATAACAAACAGCAACAACAACAGCAACAGCAACAGATTGAGAGTTCTAGTTATAACAATACTACAACGACTACTTCTGGCTCCTATAAACTTTTCCTATTCTATTTAAATTCCCTTATCCTTTTGATTGGGATTATCAGATCAGGTTGTTACCTAAATTACAATTTGGGCCCATTGAATAGCCTGTCATTTGTTTTCACTGGTTGGTATGATGGTAGTAGCTTTATTTCTAGCGATGTTACGAACGGTTTCAAATGCATTCTATATGCCTTGGTAGAAATATCTTTAGGTTTTCAAGTTTACGTAATGTTTAAAACAAGCAACTTGAAAATATGGGGTATAATGGCCAGTTTGTTGAGTATAGGCCTAGGCCTGATCGTGGTCGCGTTTCAAATCAATCTAACGATCTTGTCTCATATAAGGTTCTCTAGGGCTATCTCTACGAATAGATCCGAAGAAGAGAGCTCATTGTCTTCTGATAGCGTGGGTTATGTAATAAATAGCATTTGGATGGATCTGCCAACGATTTTATTTTCCATCAGCATAAATATCATGACAATCTTACTAATTGGTAAGCTAATCATCGCAATTAGAACAAGGAGGTACCTGGGTTTGAAACAGTTCGACTCATTTCACATCTTGTTGATTGGCTTTAGTCAAACGTTAATAATCCCGTCCATCATATTAGTTGTCCACTATTTCTATTTGTCTCAGAACAAGGATAGCTTACTTCAGCAAATTTCCTTATTGTTAATCATATTGATGTTACCGTTATCTTCATTATGGGCACAAACAGCAAACAACACCCATAATATCAACTCTTCTCCCTCACTGTCTTTTATTTCAAGGCATCATTCTTTCGACAGTTCCAGATCCGGAGGTAGTAACACTATTGTGTCCAACGGTGGTAGTAATGGTGGTGGCGGTGGTGGTGGCAACTTCCCAGTAAGTGGTATAGACGCGCAGTTACCGCCTGACATTGAGAAGATCTTACATGAAGATAATAACTATAAACTGCTTAACAGCAATAACGAGTCAGTTAACGACGGCGATATCATTATTAATGACGAGGGGATGATCACTAAGCAGATCACGATTAAAAGAGTTTAAgtcgacggatccactagtggtcatagctgtttcctg
Candida parapsilosis with CA C-terminal
gtaaaacgacggccagtctcgagaagcttTACAAAATGAATAAGATAGTCTCAAAACTTTCTTCTAGTGACGTTATAGTCACAGTAACCATTCCAAACGAAGAAGACGGTACTTATGAAGTACCATTCTATGCTATTGATAATTACCATTACAGTCGTATGGAGAACGCAGTGGTGTTGGGTGCCACGATTGGTGCTTGTTCTATGTTGTTAATCATGTTAATCGGGATTCTATTCAAGAATTTTCAAAGGTTAAGGAAATCTCTATTGTTCAACATAAACTTCGCAATTTTATTGATGCTAATTCTGAGATCTGCGTGTTATATTAACTATCTAATGAATAATCTGAGCTCTATATCATTCTTTTTCACCGGTATTTTTGACGATGAGTCATTCATGAGCTCCGATGCCGCAAATGCATTCAAAGTGATCTTAGTAGCATTAATTGAAGTTTCTTTAACCTACCAAATATACGTAATGTTTAAGACCCCCATGTTGAAGTCTTGGGGTATCTTTGCTAGTGTCTTAGCAGGTGTTTTAGGACTTGCAACGTTGGCAACACAAATATATACGACTGTAATGTCTCATGTAAATTTTGTGAACGGCACAACAGGTTCACCTTCACAGGTAACAAGTGCATGGATGGATATGCCAACAATCTTATTTTCTGTTTCTATTAACGTTTTATCTATGTTTTTGGTATGTAAGTTGGGTTTGGCCATTAGAACTAGAAGGTACTTAGGTTTAAAGCAATTCGATGCCTTCCATATTTTATTCATTATGTCAACTCAGACAATGATTATTCCTTCCATAATTTTGTTCGTCCATTACTTCGACCAGAATGATTCACAGACAACCTTGGTTAACATCAGTTTATTGTTAGTGGTCATTAGTTTGCCGTTAAGCTCTTTATGGGCACAAACAGCAAACAACACCCATAATATCAACTCTTCTCCCTCACTGTCTTTTATTTCAAGGCATCATTCTTTCGACAGTTCCAGATCCGGAGGTAGTAACACTATTGTGTCCAACGGTGGTAGTAATGGTGGTGGCGGTGGTGGTGGCAACTTCCCAGTAAGTGGTATAGACGCGCAGTTACCGCCTGACATTGAGAAGATCTTACATGAAGATAATAACTATAAACTGCTTAACAGCAATAACGAGTCAGTTAACGACGGCGATATCATTATTAATGACGAGGGGATGATCACTAAGCAGATCACGATTAAAAGAGTTTAAgtcgacggatccactagtggtcatagctgtttcctg
Candida parapsilosis with SC C-terminal
gtaaaacgacggccagtctcgagaagcttTACAAAATGAATAAGATAGTCTCAAAACTTTCTTCTAGTGACGTTATAGTCACAGTAACCATTCCAAACGAAGAAGACGGTACTTATGAAGTACCATTCTATGCTATTGATAATTACCATTACAGTCGTATGGAGAACGCAGTGGTGTTGGGTGCCACGATTGGTGCTTGTTCTATGTTGTTAATCATGTTAATCGGGATTCTATTCAAGAATTTTCAAAGGTTAAGGAAATCTCTATTGTTCAACATAAACTTCGCAATTTTATTGATGCTAATTCTGAGATCTGCGTGTTATATTAACTATCTAATGAATAATCTGAGCTCTATATCATTCTTTTTCACCGGTATTTTTGACGATGAGTCATTCATGAGCTCCGATGCCGCAAATGCATTCAAAGTGATCTTAGTAGCATTAATTGAAGTTTCTTTAACCTACCAAATATACGTAATGTTTAAGACCCCCATGTTGAAGTCTTGGGGTATCTTTGCTAGTGTCTTAGCAGGTGTTTTAGGACTTGCAACGTTGGCAACACAAATATATACGACTGTAATGTCTCATGTAAATTTTGTGAACGGCACAACAGGTTCACCTTCACAGGTAACAAGTGCATGGATGGATATGCCAACAATCTTATTTTCTGTTTCTATTAACGTTTTATCTATGTTTTTGGTATGTAAGTTGGGTTTGGCCATTAGAACTAGAAGGTACTTAGGTTTAAAGCAATTCGATGCCTTCCATATTTTATTCATTATGTCAACTCAGACAATGATTATTCCTTCCATAATTTTGTTCGTCCATTACTTCGACCAGAATGATTCACAGACAACCTTGGTTAACATCAGTTTATTGTTAGTGGTCATTAGTTTGCCGTTAAGCTCTTTATGGGCCACGGCTGCTAATAATGCATCCAAAACAAACACAATTACTTCAGACTTTACAACATCCACAGATAGGTTTTATCCAGGCACGCTGTCTAGCTTTCAAACTGATAGTATCAACAACGATGCTAAAAGCAGTCTCAGAAGTAGATTATATGACCTATATCCTAGAAGGAAGGAAACAACATCGGATAAACATTCGGAAAGAACTTTTGTTTCTGAGACTGCAGATGATATAGAGAAAAATCAGTTTTATCAGTTGCCCACACCTACGAGTTCAAAAAATACTAGGATAGGACCGTTTGCTGATGCAAGTTACAAAGAGGGAGAAGTTGAACCCGTTGACATGTACACTCCCGATACGGCAGCTGATGAGGAAGCCAGAAAGTTCTGGACTGAGGATAATAATAATTTATGAgtcgacggatccactagtggtcatagctgtttcctg
Candida tropicalis with CA C-terminal
gtaaaacgacggccagtctcgagaagcttTACAAAATGGACATTAATAACACGATTCAATCTTCCGGCGATATAATCATAACATATACTATTCCAGGTATAGAAGAACCTTTTGAACTACCCTTTGAAGTCTTGAACCATTTTCAATCTGAGCAGTCCAAAAACTGTTTGGTTATGGGAGTTATGATAGGTTCTTGCTCAGTTTTGCTTATCTTCCTGGTAGGGATCTTATTCAAGACTAATAAGTTCTCCACCATAGGTAAGAGTAAGAACTTGTCAAAGAACTTTCTTTTCTATTTGAATTGCTTGATAACATTCATAGGTATTATCAGAGCTGCGTGCTTTTCCAACTATCTTTTAGGCCCATTAAATTCCGCCAGTTTTGCCTTTACAGGCTGGTATAATGGTGAATCTTATGCTAGTTCTGAAGCTGCTAACGGTTTCCGTGTTATCTTGTTCGCATTAATTGAGACTTCAATGGTTTTTCAGGTTTTTGTAATGTTCAGGGGTGCTGGCATGAAAAAACTGGCATATTCCGTGACTATACTGTGTACTGCTTTGGCTTTAGTGGTTGTGGGTTTCCAAATTAATTCAGCTGTTCTTTCTCACAGAAGGTTTGTCAACACCGTTAATGAAATTGGTGACACTGGCTTGTCTTCTATTTGGCTTGATCTACCGACCATACTATTTAGCGTATCTGTCAATTTAATGTCTGTACTTTTGATTGGCAAGTTGATTATGGCAATAAAAACCAGAAGGTATTTGGGTCTAAAACAATTTGACAGCTTCCATGTTCTACTGATTTGCAGCACACAAACCCTTTTGGTGCCTAGTTTGATATTGTTCGTACATTACTTCCTATTTTTTAGAAATGCAAATGTGATGCTGATCAATATTTCCATTTTACTTATTGTTTTGATGCTGCCTTTTTCAAGTCTGTGGGCACAAACAGCAAACAACACCCATAATATCAACTCTTCTCCCTCACTGTCTTTTATTTCAAGGCATCATTCTTTCGACAGTTCCAGATCCGGAGGTAGTAACACTATTGTGTCCAACGGTGGTAGTAATGGTGGTGGCGGTGGTGGTGGCAACTTCCCAGTAAGTGGTATAGACGCGCAGTTACCGCCTGACATTGAGAAGATCTTACATGAAGATAATAACTATAAACTGCTTAACAGCAATAACGAGTCAGTTAACGACGGCGATATCATTATTAATGACGAGGGGATGATCACTAAGCAGATCACGATTAAAAGAGTTTAAgtcgacggatccactagtggtcatagctgtttcctg
Lodderomyces elongisporus with SC C-terminal
gtaaaacgacggccagtctcgagaagcttTACAAAATGGATGAAGCAATAAATGCCAATCTGGTCTCCGGGGATATCATTGTTTCATTCAATATACCTGGATTACCAGAGCCTGTTCAAGTGCCATTTTCAGAGTTCGACTCCTTTCATAAGGATCAATTGATTGGAGTTATTATATTAGGCGTCACTATCGGTGCTTGCTCTTTGTTGTTAATACTATTGCTTGGAATGTTGTATAAGTCACGTGAAAAGTATTGGAAGAGCTTATTGTTTATGTTGAATGTTTGTATACTTGCCGCGACAATCTTACGTTCTGGTTGCTTTTTGGATTATTATTTGTCTGATCTAGCTTCTATCAGTTATACCTTTACCGGTGTTTATAACGGCACAAGTTTTGCAAGTAGTGATGCAGCCAATGTTTTCAAGACTATTATGTTTGCACTAATAGAGACGAGTTTGACCTTTCAAGTTTATGTTATGTTCCAAGGCACCACATGGAAGAACTGGGGACATGCAGTAACCGCATTGTCCGGTTTGTTGTCAGTCGCGTCAGTAGCATTCCAGATATATACTACTATATTAAGTCACAATAACTTCAACGCAACAATTTCAGGGACTGGTACTTTAACATCTGGGGTGTGGATGGACCTGCCAACTTTACTATTTGCTGCATCAATCAATTTTATGACTATCCTACTGCTATTCAAACTTGGCATGGCTATCAGACAAAGGAGATACTTAGGTTTGAAACAGTTTGATGGCTTTCACATCCTTTTTATTATGTTTACGCAGACCTTATTTATTCCCTCTATCTTATTGGTGATCCATTATTTTTATCAAGCTATGTCTGGTCCATTCATCATAAACATGGCCCTGTTCCTTGTGGTAGCCTTCTTGCCATTGTCATCATTGTGGGCCACGGCTGCTAATAATGCATCCAAAACAAACACAATTACTTCAGACTTTACAACATCCACAGATAGGTTTTATCCAGGCACGCTGTCTAGCTTTCAAACTGATAGTATCAACAACGATGCTAAAAGCAGTCTCAGAAGTAGATTATATGACCTATATCCTAGAAGGAAGGAAACAACATCGGATAAACATTCGGAAAGAACTTTTGTTTCTGAGACTGCAGATGATATAGAGAAAAATCAGTTTTATCAGTTGCCCACACCTACGAGTTCAAAAAATACTAGGATAGGACCGTTTGCTGATGCAAGTTACAAAGAGGGAGAAGTTGAACCCGTTGACATGTACACTCCCGATACGGCAGCTGATGAGGAAGCCAGAAAGTTCTGGACTGAGGATAATAATAATTTATGAgtcgacggatccactagtggtcatagctgtttcctg
Signals
Genomic PCR of MF(ALPHA)1 ([http://www.yeastgenome.org/locus/mf%28alpha%291/overview YPL187W]) - secretion tag (first PCR), second PCR to add the actual pheromone and stop codon
TACAAAATGAGATTTCCTTCAATTTTTACTGCAGTTTTATTCGCAGCATCCTCCGCATTAGCTGCTCCAGTCAACACTACAACAGAAGATGAAACGGCACAAATTCCGGCTGAAGCTGTCATCGGTTACTTAGATTTAGAAGGGGATTTCGATGTTGCTGTTTTGCCATTTTCCAACAGCACAAATAACGGGTTATTGTTTATAAATACTACTATTGCCAGCATTGCTGCTAAAGAAGAAGGGGTATCTTTGGATAAAAGAGAGGCTGAAGCTTGGCATTGGTTGCAACTAAAACCTGGCCAACCAATGTACTAA
Primers
| Name | Sequence |
|---|---|
| ig2-AlphaP-CA-F | ggatatttcgaacctggttaagaattcGGAGCTCCAGCTTTTGTTCCC |
| ig2-AlphaP-CA-R | aaaatttgttaaacggaagccAGCTTCAGCCTCTCTTTTATCCAAAG |
| ig2-AlphaP-CP-F | ggttactatgaaccacagtaagaattcGGAGCTCCAGCTTTTGTTCCC |
| ig2-AlphaP-CP-R | gtaggttgtccaatgaggcttAGCTTCAGCCTCTCTTTTATCCAAAG |
| ig2-AlphaP-CT-F | atatggatggttttctccaaactaagaattcGGAGCTCCAGCTTTTGTTCCC |
| ig2-AlphaP-CT-R | ctggttaatctgaatttgaatttggcAGCTTCAGCCTCTCTTTTATCCAAAG |
| ig2-AlphaP-LE-F | ggaaggtttagtcctgtttaagaattcGGAGCTCCAGCTTTTGTTCCC |
| ig2-AlphaP-LE-R | atatcttgtccacatccaaccAGCTTCAGCCTCTCTTTTATCCAAAG |
Pheromones
| Name | Sequence |
|---|---|
| Saccharomyces cerevisiae | TGGCATTGGTTGCAACTAAAACCTGGCCAACCAATGTAC |
| Candida albicans | GGCTTCCGTTTAACAAATTTTGGATATTTCGAACCTGGT |
| Candida parapsilosis | AAGCCTCATTGGACAACCTACGGTTACTATGAACCACAG |
| Candida tropicalis | GCCAAATTCAAATTCAGATTAACCAGATATGGATGGTTTTCTCCAAAC |
| Lodderomyces elongisporus | GGTTGGATGTGGACAAGATATGGAAGGTTTAGTCCTGTT |
Materials and methods
Used strains
E. coli
- DH5α
S. cerevisiae
- 7283
- 7284
- Δbar1
- 6193
Used material
- LB-M agar plates with ampicillin
- 1.5 ml eppendorf tubes
- 0.5 ml PCR tubes
- 50 ml centrifuge conical base and rim tubes
- NucleoSpin Plasmid DNA, RNA, and protein purification Kit
- NucleoSpin Gel and PCR Clean-up Kit
- YPD medium, SD-min medium and minimal medium with galactosidase
- NaOH agarose gel and buffer
Used methods
- Transformation
- Miniprep
- Restriction digest
- Ligation
- NucleoSpin Gel Clean-up
- NucleoSpin Plasmid DNA purification
- Flow cytometry
All used protocols can be found at the Protocols page
Used software
- CFlow Plus
- Microsoft Excel
- MATLAB®
Construction
Validation
Receptors
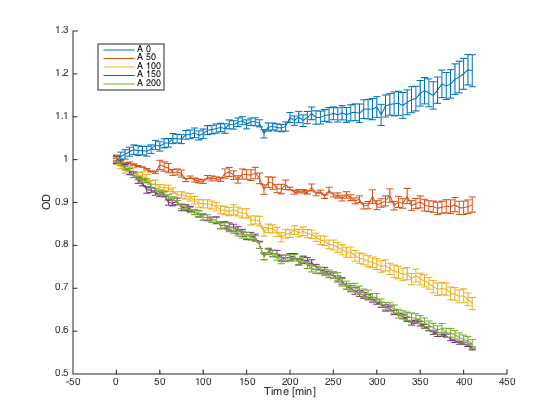
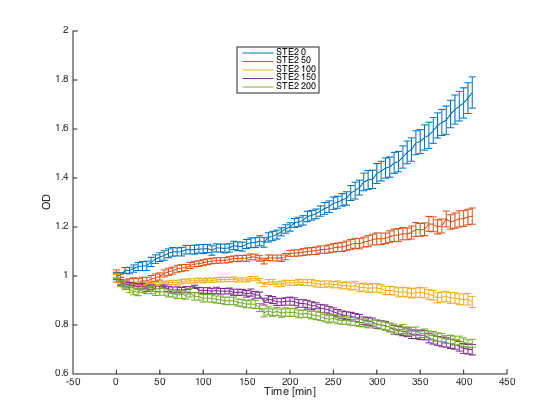
Wild-type a cells (7284) and alpha cells with pGAL1_STE2 were grown in minimal media with galactosidase overnight. At time t=0, cultures were diluted and different concentrations of the synthetic alpha pheromone were added (0, 50, 100, 150 and 200μM). In the ideal case, alpha cells with introduced STE2 receptor should exhibit similar behavior as WT a cells - growth arrest in the presence of the pheromone.
Reaction of the WT a cells to different concentrations of the pheromone:
Reaction of the alpha cells with introduced STE2 receptor to different concentration of the pheromone:
Signals
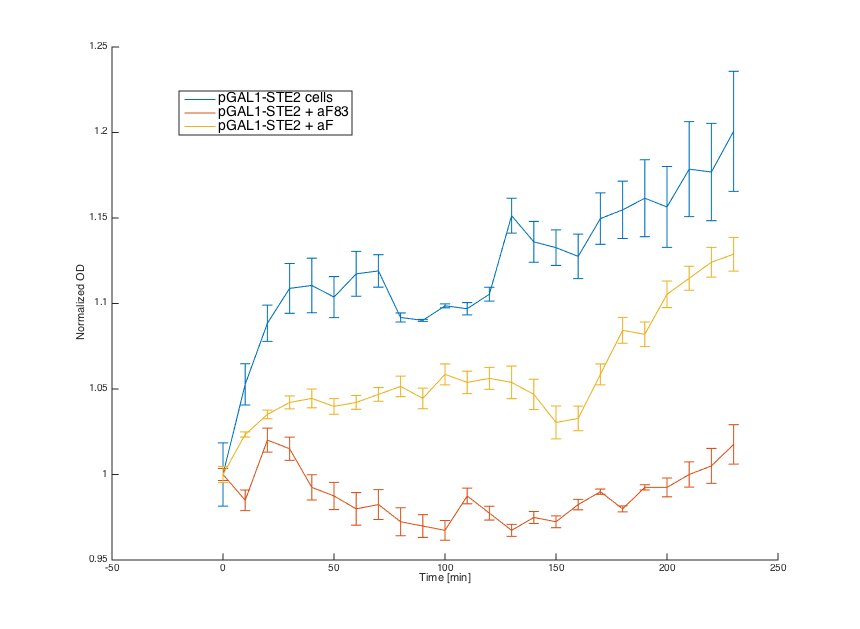
Wild-type a cells (7284) , cells with pGAL1_STE2, cells with pCUP1_Pheromone and wild-type alpha cells (7283) were grown overnight in SD min.
In the morning, cultures 7283, 7284 and pGAL1_STE2 were diluted in the same medium and incubated for 6 hours. Culture pCUP1_Pheromone was centrifuged (3000g) and resuspended in SG-min (minimal medium with galactose) supplemented with 0.1μM CuSO4.
Alpha factors for induction were obtained by centrifugation of 7283 (aF83) and pCUP1_Pheromone (aF) cultures at 3000g and removing the cell pellet. Obtained supernatant was combined with a and pGAL1_STE2 cells (20μL culture + 80μL media).
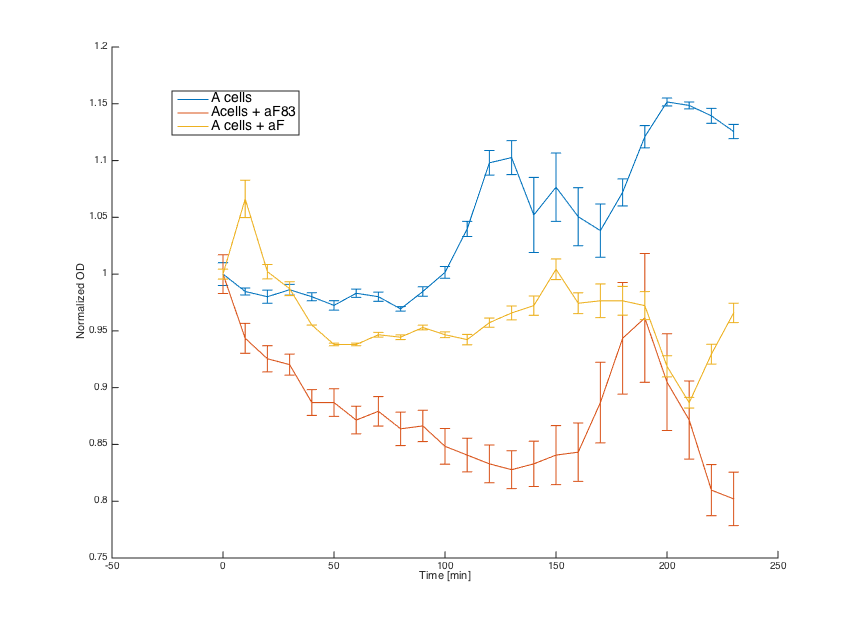
First, reaction of the a cells in the medium with WT alpha factor and the alpha factor produced by our a cells transformed with pCUP1_Pheromone was determined:
The a cells without any pheromone present exhibited normal growth after 1h from the transfer to the new medium. Cells with a WT pheromone present were arrested in their grow as expected. The same reaction was observed for the cells in the medium with pheromones produced by the a cells with our pCUP1_Pheromone plasmid, even in a smaller scale. This could be explained by a smaller production of the pheromone from the partially induced pCUP1 promoter.
Then, we tested reaction of the alpha cells transformed with pGAL1_STE2 plasmid in both media:
Cells in media without any pheromone exhibited normal growth after 1h from the transfer to the new medium. Cells in the media with WT pheromone exhibited growth arrest as expected and the cellls in media with pheromones produced by a cells with our pCUP1_Pheromone plasmid exhibited only partial arrest, which corresponds to the lower production of the pheromone (as mentioned above).
Summary: Coupling of the introduced STE2 receptor to the pheromone mating pathway and successful production and secretion of the introduced mating pheromone to a cells were demonstrated.
Results
Test of orthogonality (growth arrest approach)
Test of orthogonality (fluorescence approach)
Final constructs
References
- ↑ Lin, C.-H., Choi, a., & Bennett, R. J. (2011). Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species. Molecular Biology of the Cell, 22(24), 4918–4930. doi:10.1091/mbc.E11-09-0749
- ↑ Caplan, S., Green, R., Rocco, J., & Kurjan, J. (1991). Glycosylation and structure of the yeast MF alpha 1 alpha-factor precursor is important for efficient transport through the secretory pathway. Journal of Bacteriology, 173, 627–635. doi:10.1039/c1mb05175j
- ↑ Kurjan J, Herskowitz I. (1982) Structure of a yeast pheromone gene (MF alpha): a putative alpha-factor precursor contains four tandem copies of mature alpha-factor. Cell. 1982 Oct;30(3):933-43. doi:10.1016/0092-8674(82)90298-7
- ↑ Caplan, S., Green, R., Rocco, J., & Kurjan, J. (1991). Glycosylation and structure of the yeast MF alpha 1 alpha-factor precursor is important for efficient transport through the secretory pathway. Journal of Bacteriology, 173, 627–635. doi:10.1039/c1mb05175j
- ↑ Janiak, A. M., Sargsyan, H., Russo, J., Naider, F., Hauser, M., & Becker, J. M. (2005). Functional expression of the Candida albicans alpha-factor receptor in Saccharomyces cerevisiae. Fungal Genetics and Biology, 42(2005), 328–338. doi:10.1016/j.fgb.2005.01.006