Team:UCLA/Notebook/Spider Silk Genetics/24 July 2015
Contents
7/24/2015
Sequencing Results
- M1-AB
- All three good.
- M1-BC
- All three good.
- M1-CA
- All three good.
- M2-12(1C3)
- 1: No insert
- 2: No insert
- 3: Gibberish (does not align to anything)
- Unfortunately, no correct sequences for M2-12(1C3)
- Maybe re-transform using first ligation product.
PCR Amplification for M-12
- Amplify using post-elution primers in 1x 50 uL reaction.
| Volume (uL) | |
|---|---|
| 5x Q5 Buffer | 10 |
| 10 mM dNTPs | 1 |
| 10 uM For (F-03) | 2.5 |
| 10 uM Rev (G-03) | 2.5 |
| Template | 1 |
| Q5 Polymerase | 0.5 |
| ddH2O | 32.5 |
| Total | 50 uL |
| 98 C | 30 sec |
| 98 C | 10 sec |
| 66 C | 20 sec |
| 72 C | 40 sec |
| repeat from step 2 | 20x |
| 72 C | 2 min |
| 12 C | hold |
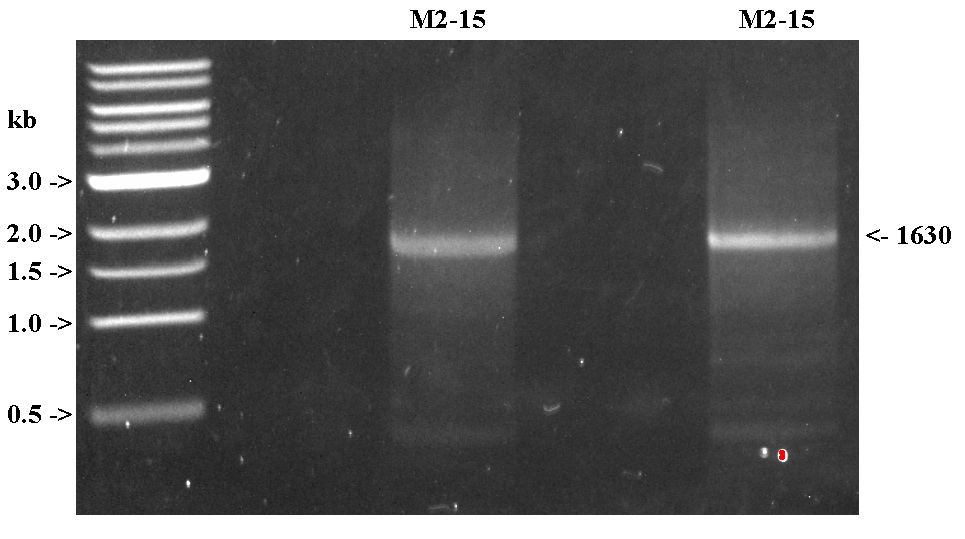
- After PCR, sample was split into two equal volumes for gel purification.
- Verified on 1% TAE gel, with 2 uL of NEB 1kb ladder. (Fasih loaded this gel, and was somewhat rough with it.).
- Not a lot of yield this time. May need to PCR more in the future.
- The 1630 bp band was excised, and gel purified using Qiagen kit.
Colony PCR for M2-12(1C3)
- Picked eight colonies from the plate of M2-12(1C3) retrieved 7/22/2015.
- Suspended colony in 15 uL LB.
- Streaked pipet tip after resuspension on an LB-Chloramphenicol index plate.
| Volume (uL) | |
|---|---|
| 2x Taq Red | 12.5 |
| 10 uM For (VF) | 1.25 |
| 10 uM Rev (VR) | 1.25 |
| Template (1:50) | 1 uL |
| ddH2O | 9 uL |
| Total | 25 uL |
| 95 C | 3 min |
| 95 C | 25 sec |
| 56 C | 30 sec |
| 72 C | 45 sec |
| repeat from step 2 | 30x |
| 72 C | 5 min |
| 12 C | hold |
- Negative control colony PCR: instead of colony, used 1 uL of plain LB.
Results
- Visualized colony PCR on 1% TAE gel, using 3 uL of NEB 1kb ladder.
- Colony PCR indicates that all of the colonies we tested do not have the desired insert. This is likely symptomatic of the entire plate.
- We will abandon this plate of bacteria, since it most likely does not contain any usable colonies.