Difference between revisions of "Team:Exeter/Toehold Design"
| Line 12: | Line 12: | ||
<p>NUPACK is a software package which is used in predicting (deoxy)ribonucleic acid secondary structures. It is available either as a source code which can be downloaded and run on a local computer, or it can be used online directly from the NUPACK website. NUPACK has three main functions, all of which we used in designing our Toehold sequences.</p> | <p>NUPACK is a software package which is used in predicting (deoxy)ribonucleic acid secondary structures. It is available either as a source code which can be downloaded and run on a local computer, or it can be used online directly from the NUPACK website. NUPACK has three main functions, all of which we used in designing our Toehold sequences.</p> | ||
| − | <div class="picture" style = "float: right;"> | + | <div class="picture" style = "float: right; padding-left:5px;"> |
<img src="https://static.igem.org/mediawiki/2015/7/7a/Nupack_logo.png"/> | <img src="https://static.igem.org/mediawiki/2015/7/7a/Nupack_logo.png"/> | ||
</div> | </div> | ||
Revision as of 08:48, 14 September 2015
Designing the Toeholds
"In the beginning, there were riboswitches, but Green et al. saw that they were inefficient and difficult to engineer, and so they designed Toehold Switches. Exeter iGem saw these Toeholds, and they saw that they were good, and so they decided to standardise and optimise them for use in diagnostic testing..."
NUPACK - NUcleic acid PACKage:
NUPACK is a software package which is used in predicting (deoxy)ribonucleic acid secondary structures. It is available either as a source code which can be downloaded and run on a local computer, or it can be used online directly from the NUPACK website. NUPACK has three main functions, all of which we used in designing our Toehold sequences.

Design:
The Design page on the NUPACK website is used to find an optimal sequence for a specific RNA/DNA structure at equilibrium.
Analysis:
NUPACK can be used to analyse in silico the thermodynamics of a nucleic acid sequence. It is able to show the minimum free energy (MFE) secondary structure at equilibrium and base-pairing probabilities for either a single strand, or a complex of strands. NUPACK is also able to carry out a melt on the strands across a range of different temperatures.
Utilities:
The utilities tool of NUPACK is useful for editing and viewing the nucleic acid sequence and seeing the effect it has on the MFE secondary structure. It can also be used to annotate and view the secondary structure in different ways.
The First Design:
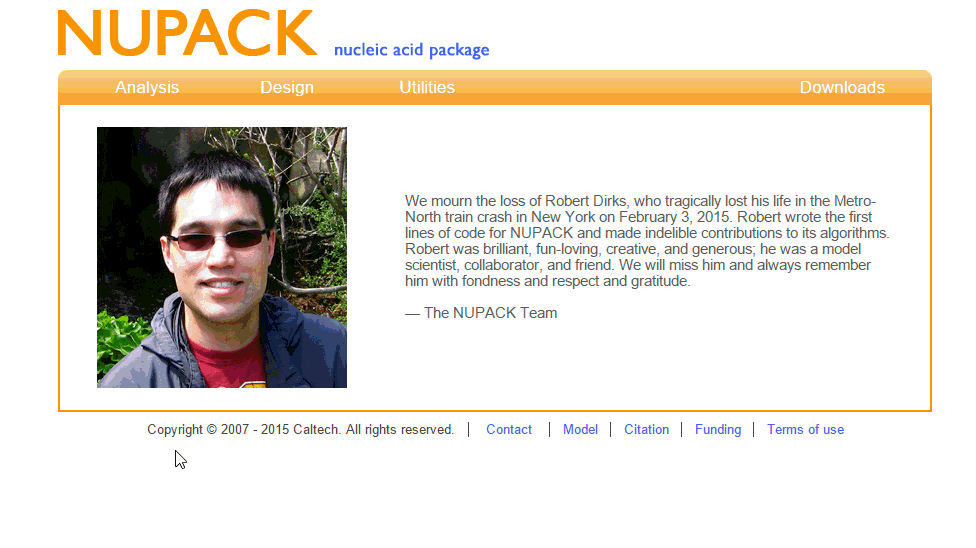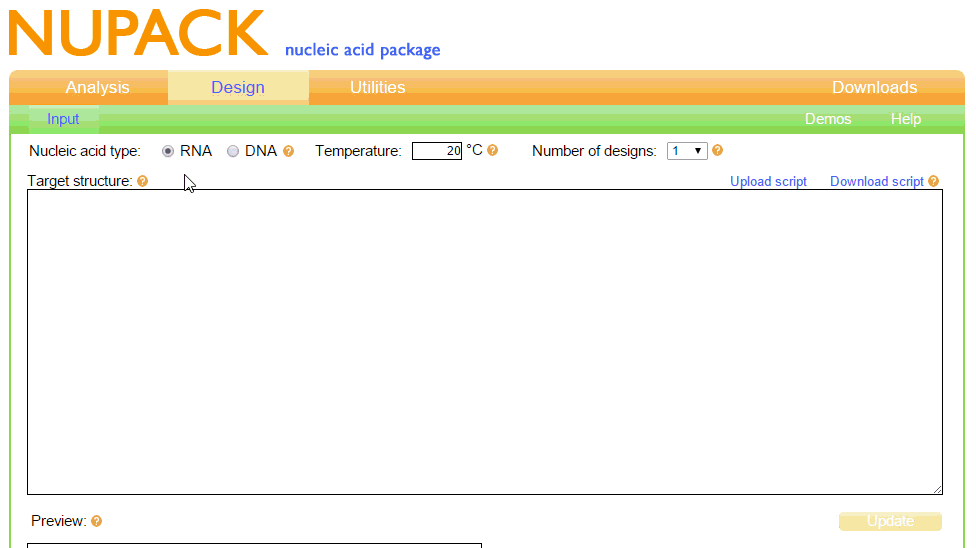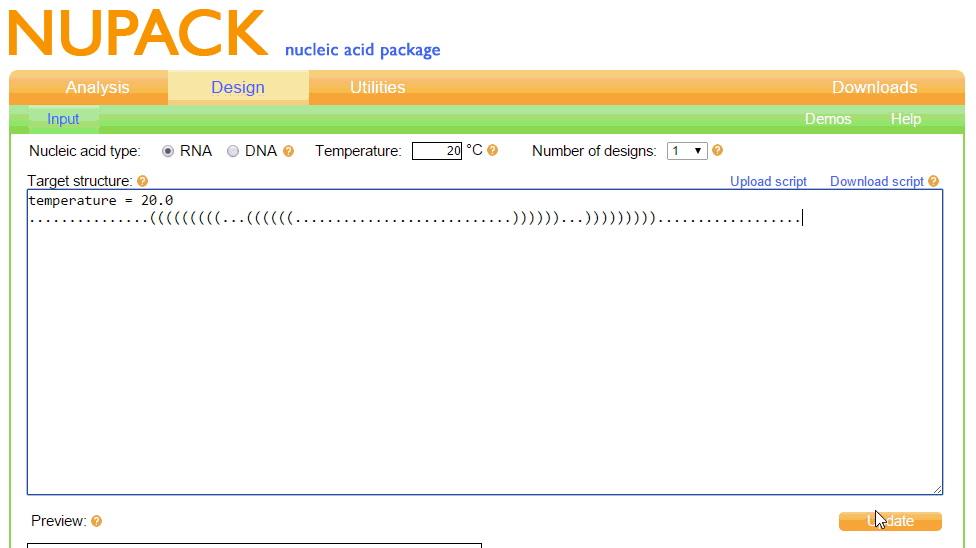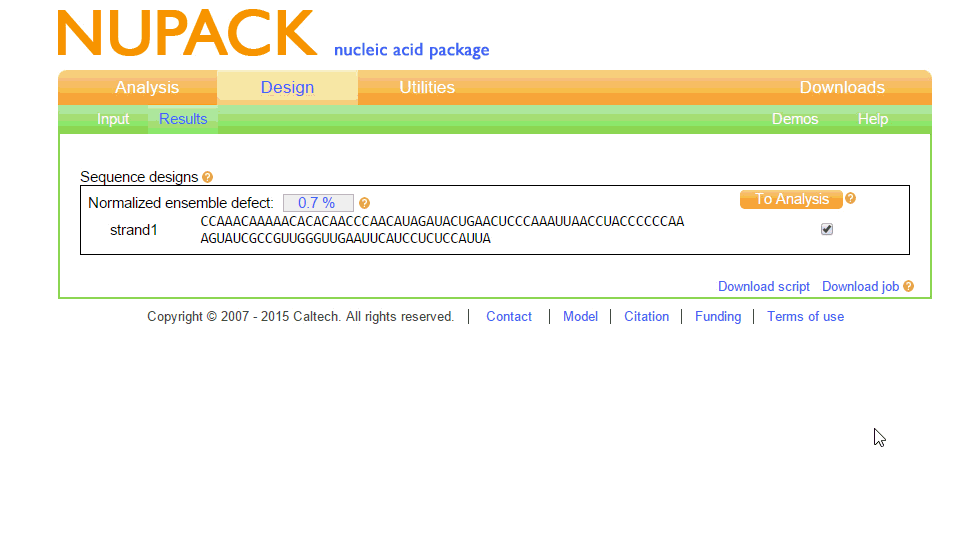
Our Toehold switches are based on the original design by Green et al. (2014). To begin, we wished to find the optimal sequence for the Toehold structure at room temperature (20C), as this is around the temperature which we anticipate our test would need to work at. To do this, we first needed to learn dot-plus-parenthesis notation, as this is how NUPACK works with structures. The notation was relatively easy to learn; dots mean unpaired bases, parenthesis mean paired bases, and pluses are used to include more than one RNA strand in the simulation, allowing RNA complexes to be formed. The input and output of this from NUPACK is shown in figure 1.