Team:Exeter/Toehold Design
Designing the Toeholds
NUPACK - NUcleic acid PACKage:
NUPACK is a software package which is used in predicting (deoxy)ribonucleic acid secondary structures. It is available either as a source code which can be downloaded and run on a local computer, or it can be used online directly from the NUPACK website. NUPACK has three main functions, all of which we used in designing our Toehold sequences.
Design:
The Design page on the NUPACK website is used to find an optimal sequence for a specific RNA/DNA structure at equilibrium.
Analysis:
NUPACK can be used to analyse in silico the thermodynamics of a nucleic acid sequence. It is able to show the minimum free energy (MFE) secondary structure at equilibrium and base-pairing probabilities for either a single strand, or a complex of strands. NUPACK is also able to carry out a melt on the strands across a range of different temperatures.
Utilities:
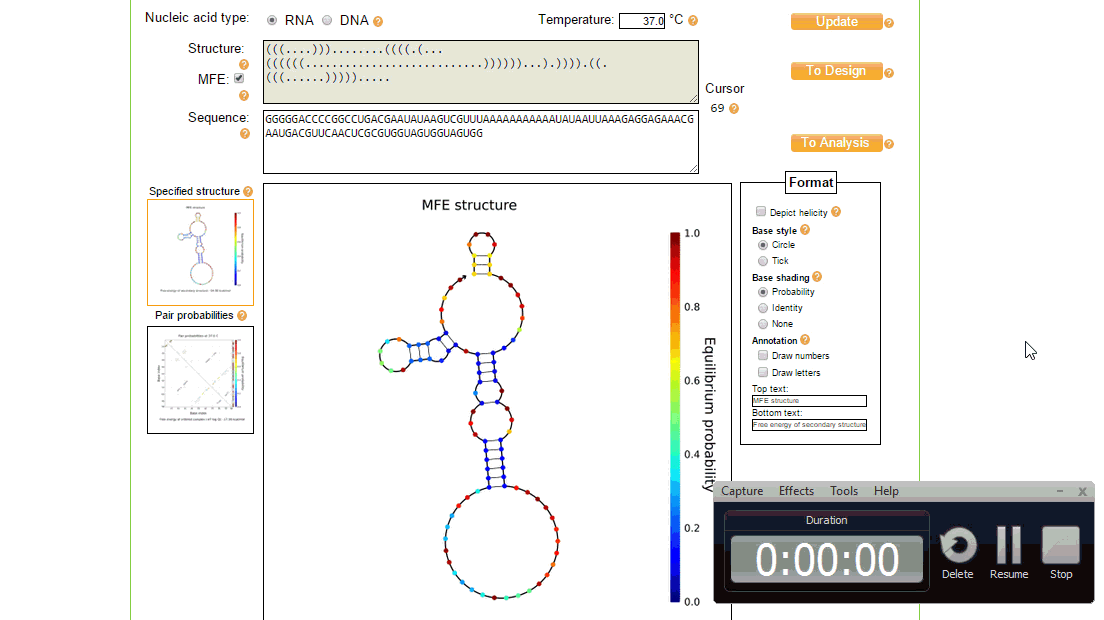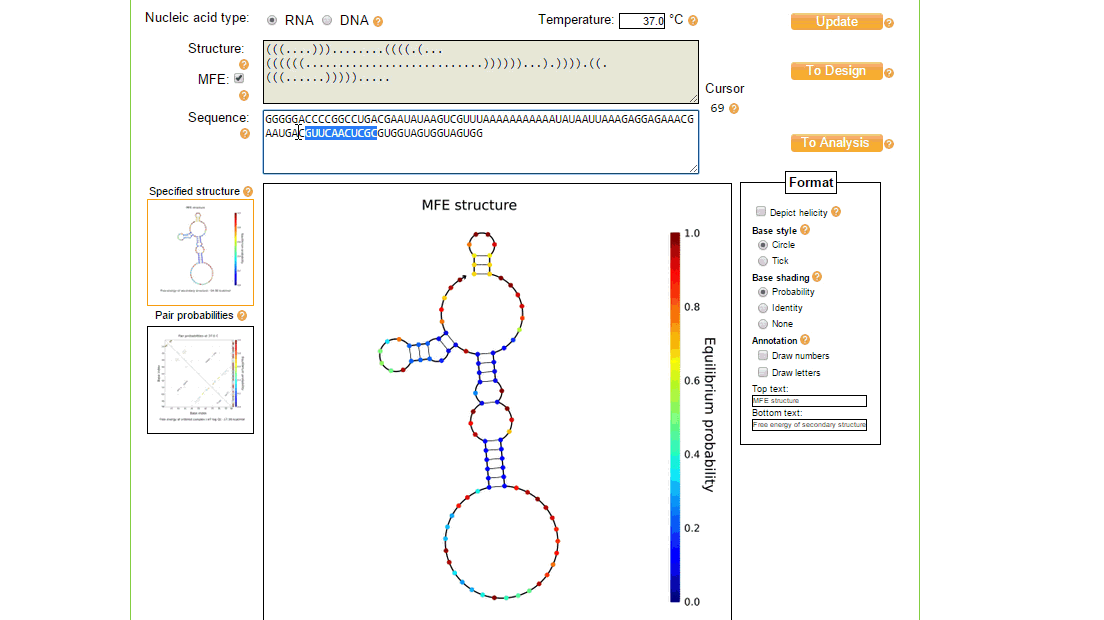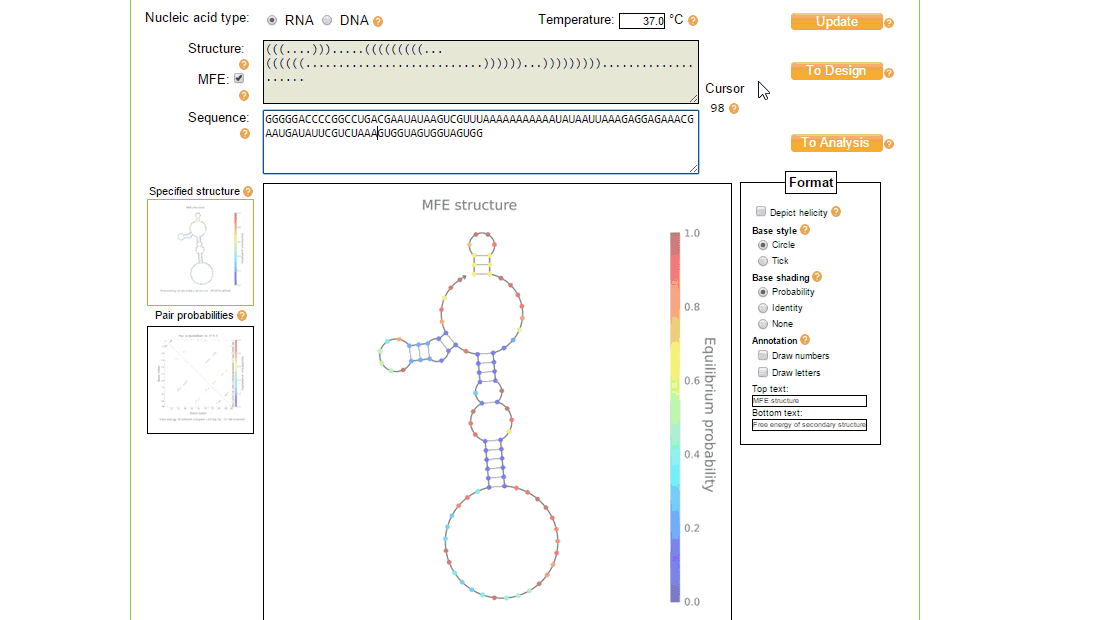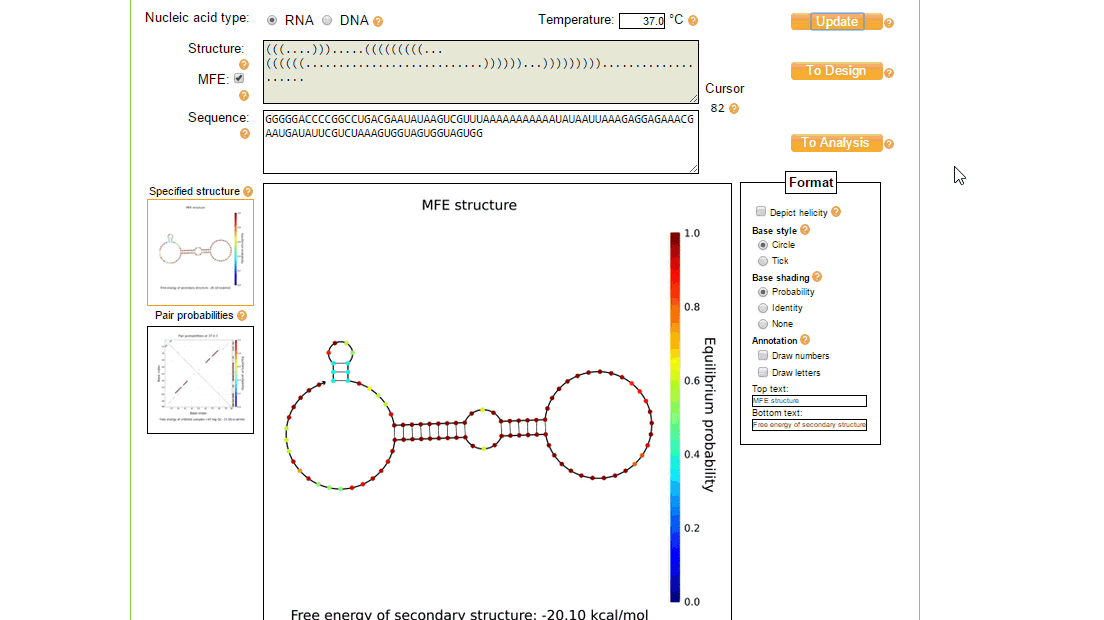
The utilities tool of NUPACK is useful for editing and viewing the nucleic acid sequence and seeing the effect it has on the MFE secondary structure. It can also be used to annotate and view the secondary structure in different ways.
The structure:
Functionality:
In order for the toehold switch to be functional, there are three main regions which must be included; the switch, the RBS, and the start codon/linker/protein coding region (figure 2). In order for the toehold switch to work, these regions must not only be present, but must stick to some important design constraints.
The Switch region:
The switch region is the part of the toehold which recognises the trigger RNA, and therefore must contain a sequence complementary to that of a section of the trigger to allow binding. In addition to this, the first section of the switch (the switch front; ~12 nucleotides not including the GGG leader sequence) must be unbound from the rest of the toehold structure to allow the trigger to easily bind if present. For this to happen, a constraint is imposed that the switch front and linker end must not be complementary. As well as the switch front being available to bind to the trigger, it is equally important that the rest of the switch region is complementary to the region before the linker end. This complementation allows a stem to be formed, which then allows the loop formation at the top. This loop region is important in ensuring that when the trigger RNA is not bound, the toehold is 'off', i.e. the protein coding region is not translated.
The ribosome binding site (RBS):
The RBS site must be located either wholly or mostly within the loop structure of the toehold to ensure that it is sequestered away from ribosomes (when the toehold is inactive), hence preventing ribosomes from binding and translating the protein coding region. Briefly, the positioning of the RBS in the loop stops ribosomes from binding as linear-linear binding of ribosome to RBS is much more favourable than loop-linear binding, which is what would have to occur when the RBS is within the loop. When the RBS is inserted into the loop, it is important to ensure that the loop structure remains. To ensure this, the loop extensions region must not have any complementary regions to the RBS.
AUG start codon/linker/protein coding region:
The AUG start codon can be found not too far from the RBS sequence. On the images shown, the AUG codon is within a mini-loop, however this mini loop does not appear to have any effect on the functionality of the toehold switch. This means that a constraint on the switch region is loosened slightly as the part of the switch which lines up with the AUG start codon in the stem does not have to be complementary to AUG, but neither does it have to be ensured that the region is not complementary.
Other requirements:
As well as the consideration above, there are a couple of other requirements to allow full toehold functionality. The first of these is the GGG leader sequence preceding the switch region. The purpose of this GGG leader sequence is not actually to do with the function of the toehold, but to ensure efficient transcription of the RNA toehold from plasmid DNA. The other important requirements are to do with the main linker. As the AUG start codon is positioned before the actual protein coding region, it is important that the linker (whose purpose is to ensure that the toehold structure remains intact) does not interfere too much with the amino acid sequence produced. The first way in which this is ensured is by making the linker region length a multiple of three so that the protein coding region remains in-frame. The second constraint on the linker is that it must not contain an in-frame stop codon, as this would result in a truncated protein being produced.
NUPACK utilities
In order to implement these changes, from the initial design page, the sequence was loaded in the utilities page where the structure and sequence can be viewed together and modified, as shown in figure 3. The process of creating a new design of toehold can take a while and several iterations to ensure full functionality, however once the toehold has been made the generic sequence can be modified to both target different trigger RNAs (by changing the switch and stem regions so that the design considerations above still match), and contain a different protein coding region to, for example, express a different type of reporter.