Laboratory Notebook
The promoter library was the first subproject that we began to approach in the lab. The work started in late april.
15-04-22
Biotec lab; Attendens: Sven, Sarah, Tobi
consumable material:
Made liquide [http://igem.rwth-aachen.de/2015/index.php/Manual/LB_Medium LB-Media] and LB-Agarplates with AmpR.
Topic Promotor Library:
Primer #XLMF#, #NNF3#, #BXT8# diluted to stock in Box P.
Primer #XLMF#, #NNF3#, #BXT8# diluted to work with in Box P'.
Suspend J23100-J23105 in pSB_J61002 from Kit Plate 4 (2014).
[http://igem.rwth-aachen.de/2015/index.php/Manual/transformation Transformation] of J23100-J23105 in pSB_J61002 into competend DH5α (from Schwaneberg)
The transformants incubate in plates in the 37 °C since 5 pm.
Next steps for tomorrow:
Make overnight cultures of J23100-J23105 in pSB_J61002.
Make LB-Media with Chloramphenicol (plates and liquids).
suspend the other Promotors from Kit4 2014 and transform them.
15-04-23
Biotec lab; Attendens: Sven, Sarah, Tobi
consumable material:
made 3x 1 ml of Chloramphenicol antibiotic for stock.
LB-Media with chloramphenicol for plates. => plates are in 4 °C fridge.
Promotor Library
J23106,J23107, J23110, J23113-18 are [http://igem.rwth-aachen.de/2015/index.php/Manual/transformation transformed] in DH5α. The plates are cultivating at 37 °C since 11:45 am.
Transformationefficiency of J23100-J23105 in pSB_J61002.
| Template |
efficiency [(cells) cfu/ng (DNA)]
|
| J23105 |
160
|
| J23104 |
650
|
| J23103 |
88
|
| J23102 |
940
|
| J23101 |
68
|
| J23100 |
41
|
J23100-J23105 in pSB_J61002 are picked from plate. 3 clones of each is in an overnight culture at 37 °C
Next steps
Plasmidprep from the overnight cultures of J23100-J23105 in pSB_J61002.
15-04-24
Biotec lab; Attendens: Sarah, Tobi, Sven
consumable material:
sterile Eppis (1.5 ml)
Agarosegel (solution in 65 °C)
Promotor Library
Plates of J23106,J23107, J23110, J23113-18 are in 4 °C fridge.
Make Cryo of J23100-J23105 in pSB_J61002 from overnight cultures (always #1). (We choose only one of the clones.)
Make Plasmidprep with rest of overnight culture from the selected clone (always #1). Measure plasmid concentration with Nanodrop.
Next steps on Monday
Look at transformation efficiency of J23106,J23107, J23110, J23113-18.
Make overnight cultures of J23106,J23107, J23110, J23113-18.
Solute from Kit and transformation of J23108, -109, -111, -119 (all in pSB1C3).
Solute from Kit and transformation of BBa_K118017 in pSB1C3.
Gradient PCR with Primer NNF3(Tm 58 °C) and BXT8(Tm 64 °C) (primers for backbone amplification). Control with gel.
Do PCR for backbone amplification.
15-04-27
Biotec lab; Attendens: Sven, Laura G., Caro, Jonas, Tobi
consumable material:
Promotor Library
Transformation efficiency of J23106, -07, -10, -13-18 in pSB_J61002 (incubation time was 20h instead of 12h).
| Template |
efficiency [(cells) cfu/ng (DNA)]
|
| J23106 |
1200
|
| J23107 |
5200
|
| J23110 |
5200
|
| J23113 |
ca 1300
|
| J23114 |
1800
|
| J23115 |
ca 3600
|
| J23116 |
2000
|
| J23117 |
3200
|
| J23118 |
1600
|
Transformation of J23108, 09, 11, 19 and BBa_K118018 (glgC+rbs) and cultivation on plates since 13:00 (accidently 1000 µl are on the plate).
Make overnight cultures of J23106,J23107, J23110, J23113-18 (We picked 2 clones of each; cultivation started at 16:20).
PCR II of J23100-J23105 in pSB_J61002 with A9W9 and XE3D (15-04-27 Gel upper row). Digested by DpnI. PCR Products are in -20 °C freezer in Falkon #1 (EOLX, VCZX, NDS4, H9MQ, T6FB, KQN8).
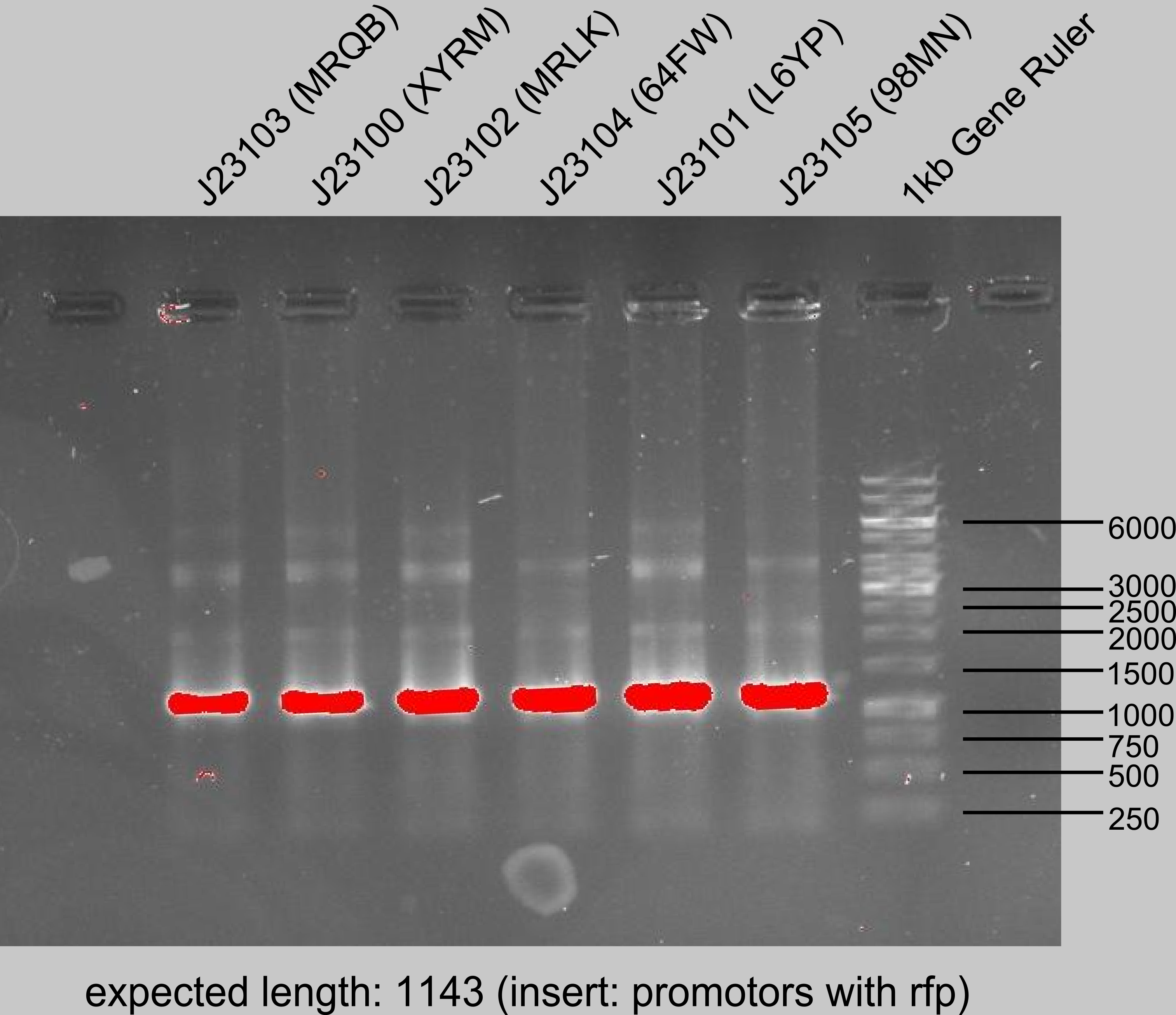
Gradient PCR with J23102 (MRLK) for primer NNF3 and BXT8 (15-04-27 Gel lower row). Must be done once again, bands did not look good!
Next steps on Tuesday
purification of PCR Products.
Gel annotation from PCR 15-04-27.
Cryo culture of J23106,J23107, J23110, J23113-18.
Plasmidprep of J23106,J23107, J23110, J23113-18.
Overnight cultures of J23108, 09, 11, 19 and BBa_K118018 (glgC+rbs).
15-04-28
Biotec lab; Attendens: Tobi, Sven, Laura G, Caro
consumable material:
sterile Eppis
Promotor Library
Evaluation: The colour of overnight cultures of J23106, J23107, J23110, J23113 - J23118 fits to their strength.
Annotation of PCR gel from promoter amplification J23100-05 (15-04-27 upper row).
Purification of the PCR products J23100-105. Measurement of concentration after digestion with DpnI and heat inactivation for 20 minutes at 80 °C
| Promoter |
Concentration (ng/ul) |
ID
|
| J23100 |
89.7 |
E0LX
|
| J23101 |
107.9 |
VYN9
|
| J23102 |
94.0 |
NDS4
|
| J23103 |
89.8 |
H9MQ
|
| J23104 |
90.4 |
T6FB
|
| J23105 |
85.0 |
KQN8
|
Cryo of J23106,J23107, J23110, J23113-18.
Plasmidprep of J23106,J23107, J23110, J23113-18. Measurement of concentrations. All in Box "Pink" row F1-9.
Transformation efficiency of J23111 in pSB1C3 is 800 cfu/ng. Of BBa_K118018 (GlgC+RBS) 36 nfu/ng (perhaps due to chloramphenicol?). All other plates "many" (keep in mind that 1 ml was plated). None of the colonies are visibly red.
Overnight cultures of J23108, 09, 11, 19 and BBa_K118018 (glgC+rbs). Started at 16:00.
Gradient PCR with J23102 (MRLK) for primer NNF3 and BXT8 (63.2-69.7 °C). Tubes are in -20 freezer to make the gel tomorrow.
Next steps
Run gel of gradient PCR.
Cryo of J23108, 09, 11, 19 and BBa_K118018 (glgC+rbs).
Plasmidprep of J23108, 09, 11, 19 and BBa_K118018 (glgC+rbs).
if time: PCR II of J23106,J23107, J23110, J23113-18 for promotoramplification (Primer A9W9 and XE3D). =>after DpnI digestion and purification with kit.
15-04-29
Biotec lab; Attendens: Tobi, Sven, Jonas, Laura G., Caro
consumable material:
Promotor Library
Overnight cultures of J23108, 09, 11 and 19 are colourless.
Cryos and plasmid prep of J23108, 09,11,19 and BBa_K118018 (GlgC+ RBS). See concentrations below.
| Part |
Concentration (ng/ul) |
ID
|
| J23108 |
49.3 |
W1KW
|
| J23109 |
44.7 |
W49P
|
| J23111 |
49.2 |
S8R6
|
| J23119 |
49.2 |
RWNO
|
| BBa_K118018 (GlgC+RBS) |
250.8 |
3Q8D
|
| BBa_K118018 (GlgC+RBS) Nr.2 |
328.7 |
V3SC
|
Gel of gradient PCR of BXT8 and NNF3 (Tip: next time always 100V)
Next steps
get gel picture from server and do annotation (from gradient PCR 15-04-29)
PCR II of J23106,J23107, J23110, J23113-18 for promotor amplification (Primer A9W9 and XE3D). =>afterwards DpnI digestion and purification with kit
PCR II of J23108, 09,11,19 for promotor amplification.
order primer and gene synthesis (first talk to Ljubica) (=> primer BXT8 might be wrong designed)
15-04-30
Biotec lab; Attendens: Jonas, Tobi, Sven, Caro
consumable material:
Promotor Library
Primers for assamblies are ordered (plus BXT8 is wrong designed).
gel picture (from gradient PCR 15-04-29) is absolutely nonsense and will not be annotated.
PCR II of J23106,J23107, J23110, J23113-14, 16-18 and J23108, 09,11,19 for promotor amplification (Primer A9W9 and XE3D), each 4 reactions per sample.
- Run the gel of PCR II from today (12 samples)
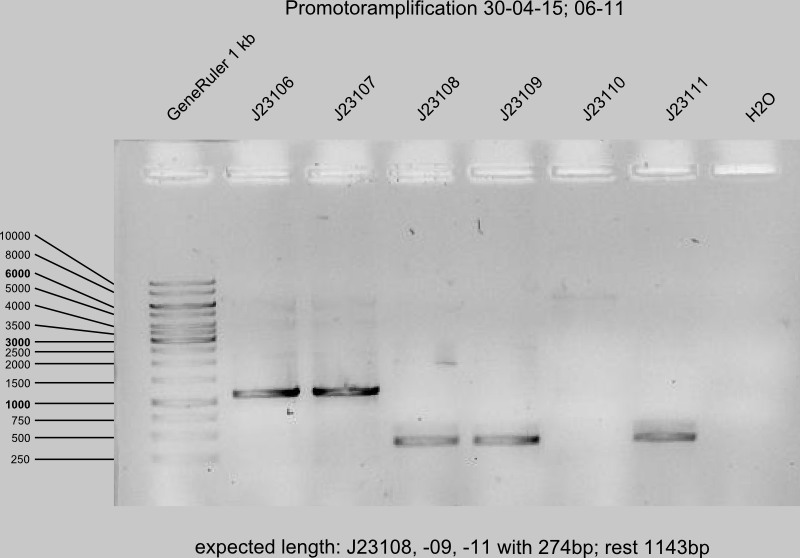
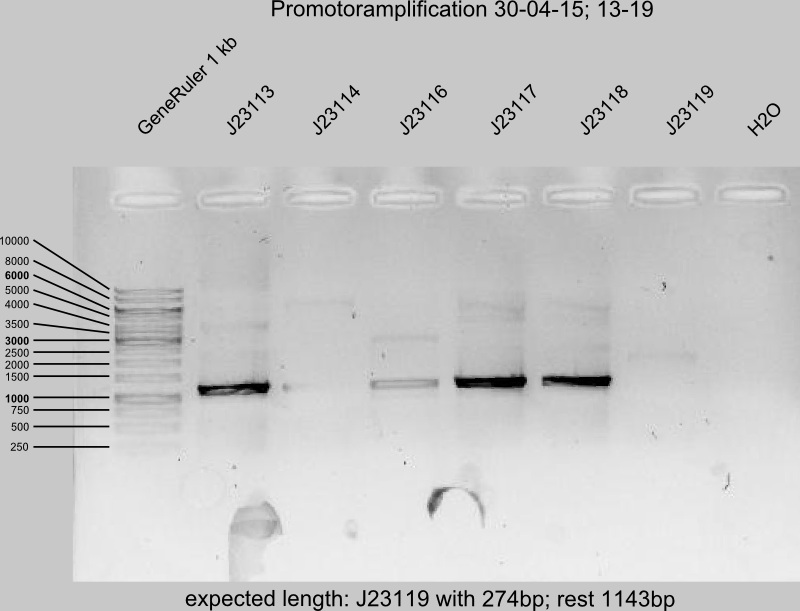
- successful: J23106, -07, -08, -09, -11, -13, -17, -18
- failed: J23110, -14, -19
- insecure: J23116
Next steps on Monday
DpnI digestion and purification of successful PCR-products (J23106, -07, -08, -09, -11, -13, -17, -18).
PCR II of J23100-05 in 4 preparations each (Primer A9W9 and XE3D). PCR of BBa_K118018 (GlgC+ RBS) (Templates: 3Q8D, V3SC) for control! (Maybe do PCR with J23116 again.)
DpnI digestion of the successful PCR-products.
Next week:
gradient PCR I with new Primers
PCR I for backbone amplification with lacI
15-05-04
Biotec lab; Attendens: Sarah, Tobi, Sven, Jonas, Laura 2x, Caro
consumable material:
Kanamycin stock 1000x in 10 Eppis (-20 °C freezer)
Promotor Library
gelcontrol of J23110, -14, -16 and -19 of all 4 preparations.
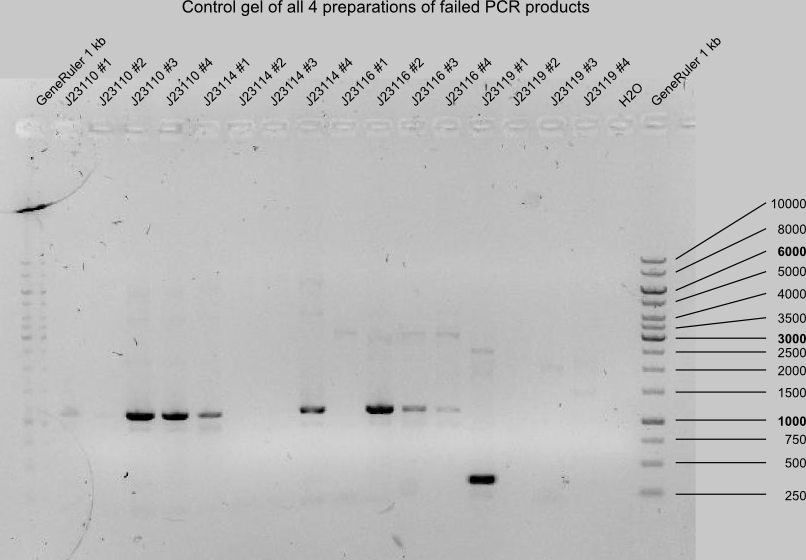
- PCR products with expected length: J23110 #3+4, J23114 #1+4, J23116 #2+3, J23119 #1
(estimated mistakes: too little buffer in PCR from 15.04-30)
- digestion of J23110, -14, -16, -19 with DpnI
DpnI digestion of successful PCR-products (J23106, -07, -08, -09, -11, -13, -17, -18) (all in separate Eppis).
PCR II of J23100-05 in 3 preparations each, -16 in 4 preparations.
=> Gel control (start: 15:30), all preparations loaded.
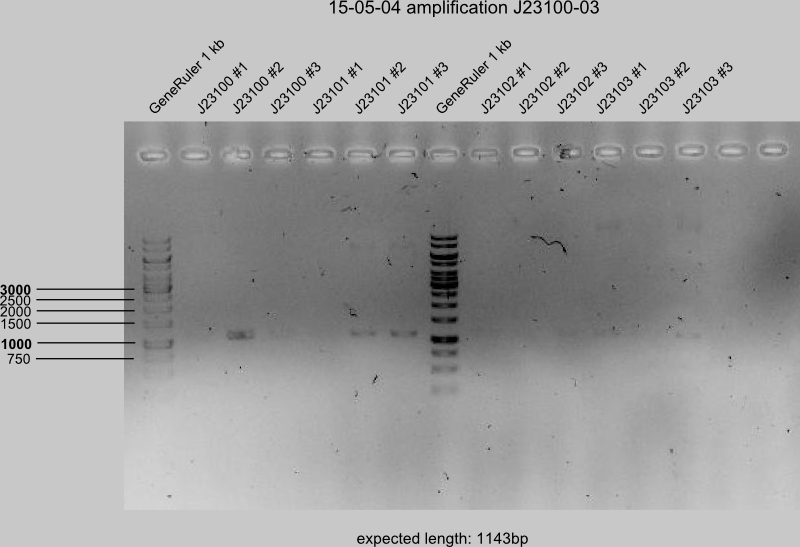
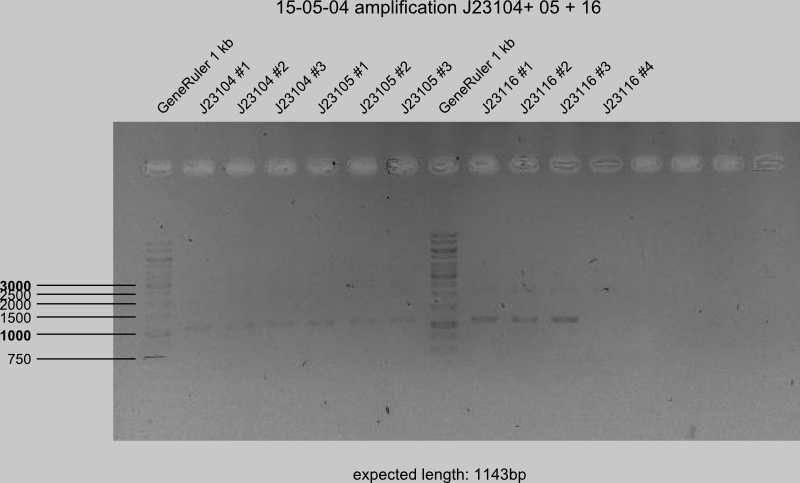
Next steps on Tuesday
Gel for recontrol of failed PCR products of J23100-05 + -16.
Heat inactivation and purification of digested J23106, -07, -08, -09, -11, -13, -17, -18.
Heat inactivation and purification of digested J23110 #3+4, J23114 #1+4, J23116 #2+3, J23119 #1. Use all these as template for new promotor-amplifications.
PCR II J23115 in 4 preparations and PCR of BBa_K118018 (GlgC+ RBS) (Templates: 3Q8D, V3SC) in 4 preparations each (for control).
=> gel for control
=> DpnI overnight of the successful PCR products.
if Primers arrived: gradient PCR I with new Primers for backbone amplification with lacI
15-05-05
Biotec lab; Attendens: Laura G., Tobi, Sven, Caro, Jonas, Michael
consumable material:
Kanamycin plates
Promotor Library
- make new gels of ugly gels from yesterday (J23100-05, -16)
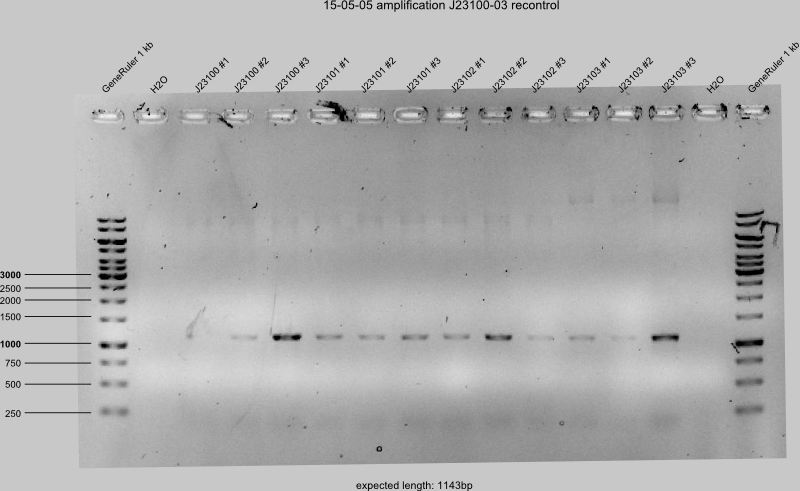
- Heat inactivation and purification of digested J23106, -07, -08, -09, -11, -13, -17, -18.
- Heat inactivation and purification of digested J23110 #3+4, J23114 #1+4, J23116 #2+3, J23119 #1
The only successful concentration measurements: J23106, -07, -09, -17 & -18. All in Pink Box with ID.
PCR II of J23108, J23110, J23111, J23113, J23114, J23115, J23116, J23119 with Plasmids from stock. Each in 3 preparations (a, b, c).
Next steps
Gel of PCR II with J23108, J23110, J23111, J23113, J23114, J23115, J23116, J23119, maybe DpnI digestion afterwards with successful PCR products (overnight).
New gradient PCR for Backboneamplification with new Primer (NNF3 & FYTO)
15-05-06
Biotec lab; Attendens: Sven, Tobi, Jonas, Laura
consumable material:
Promotor Library
Gel control of the repeated PCR with J23108, -10, -11, -13, -14, -15, -16, -19.
DpnI digestion
Successful: -08,-10,-11,-13,-14,-16,-19b (19 a,c)
Uncertain: -15 (expected length 185 bp)
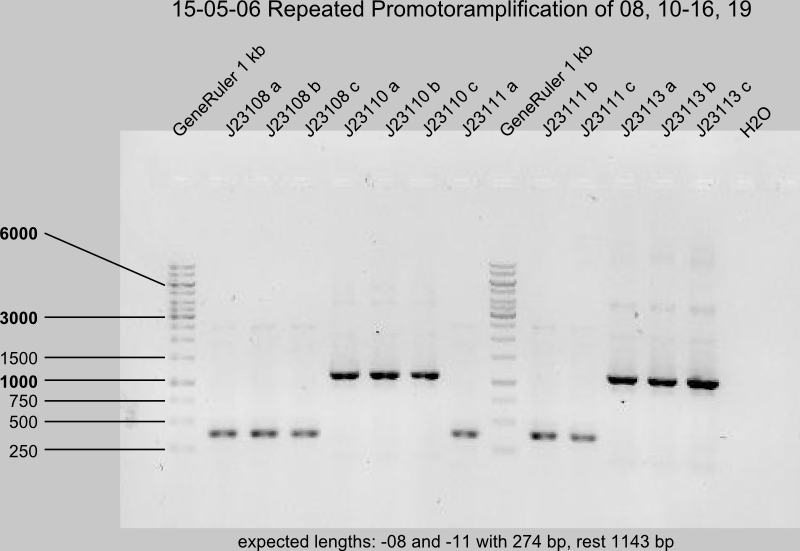
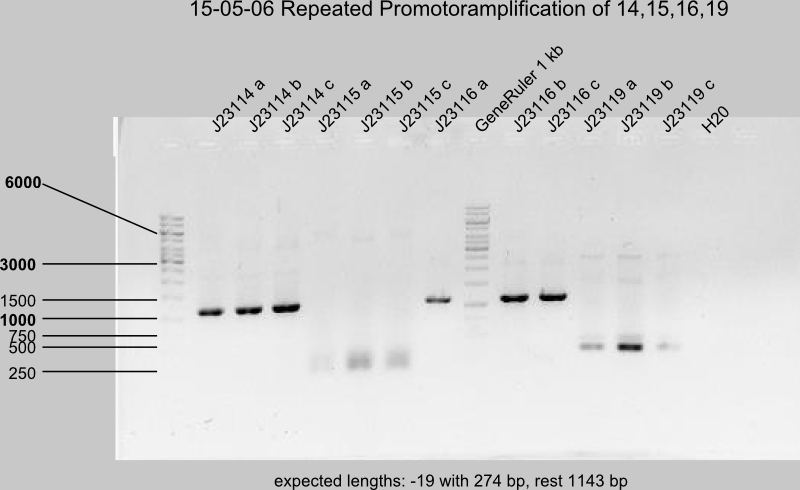
expected length of J23115 is 185 bp.
Next steps
- Purification of DpnI-digested PCR products
- Gradient PCR for backbone (PCR mix is in freezer! Has to be done on Thursday 15-05-07. PCR machine was blocked)
- Gel control for Gradient PCR
- Final PCR for backbone amplification + control
- repeat PCR with J23115
15-05-07
Biotec lab; Attendens: Sven, Tobi, Jonas, Laura,
consumable material:
2 % Agarose for gel in 65 °C
Promotor Library
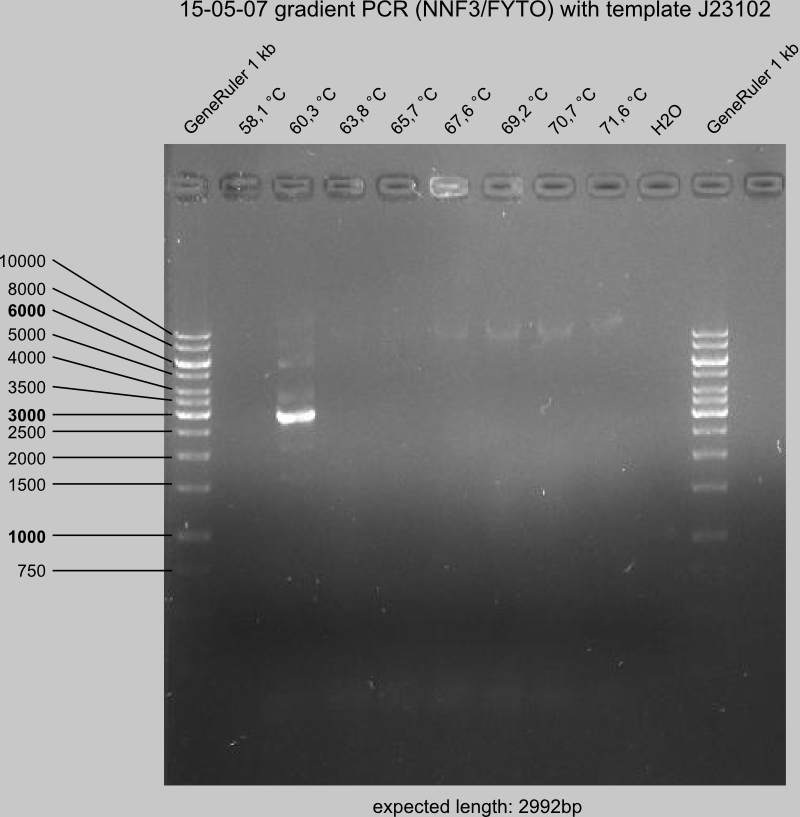
Run new gradient PCR to find out annealing temperatures for backbone amplification (primers: NNF3 and FYTO, template: MRLK)
Do the gel control of the new gradient PCR
Purification of PCR products of J23108, J23110, J23111, J23113, J23114, J23116 & J23119. All are containered and in Box "Pink".
Do 2 % Agarosegel for loading J23115 with 50 bp ladder.
Do backbone amplification with NNF3/FYTO at 60.3 °C in 10 preparations, all with same template. Run all in one slott on a gel and extract backbone from gel (purify and have a look how much is still missing).
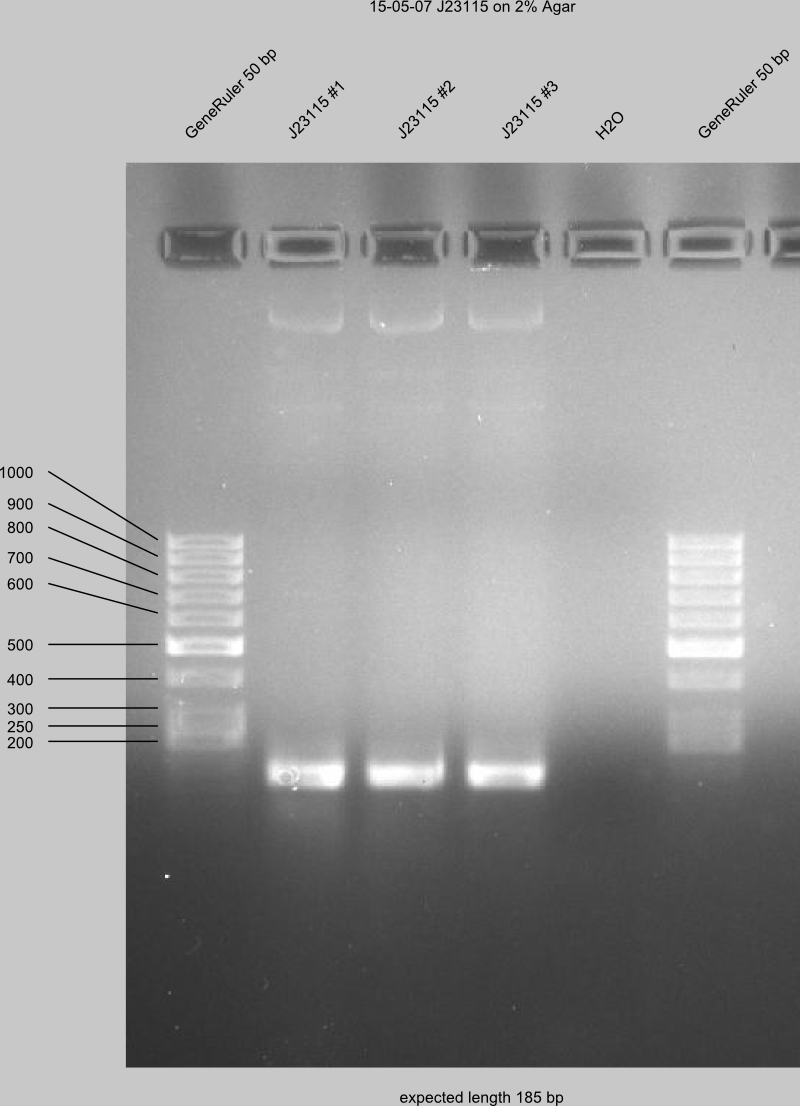
The gel of J23115 shows bands at the expected length. Therefore, digestion with DpnI since 16:30 at 37 °C.
Made PCR I for backbone amplification in 10 preparations (Is in freezer for the gel tomorrow)
Next steps
run 2 % gel of backbone (PCR I) and extract backbone (2992 bp).
do PCR I for backbone amplification again for more backbones => run gel and extract again.
purification of J23115
15-05-08
Biotec lab; Attendens: Sven, Laura G., Tobi
consumable material:
Promotor Library
purification of PCR product J23115
2 % Gel for Backbone isolation => do backbone purification with kit => 10 new containers with new "J61002 with lacI"
PCR I for more backbone => 2 % gel loaded and cutted out (and freezed over the weekend (Eppi A-H))
Next steps
purification of cutted gel with J61002 with LacI
perhaps: more amplificates of J23100-J23105
15-05-11
Biotec lab; Attendens: Laura G., Tobi, Sarah, Sven
consumable material:
petri dishes 2x
Promotor Library
We did another purification of cut agarose gel with J61002+LacI. The concentration measurement was nonsense.
It had to be checked if we already have enough backbone template for restriction and ligation.
We digested the backbone (different enzymes for "with LacI-BS" and "normal") and heat inactivated.
Next steps (in general)
[http://igem.rwth-aachen.de/2015/index.php/Manual/DigestionBB-Assembly restriction] with Xba and EcoRI of backbone (DRPV) in 10 preparations with 20 µl reaction volume each (this is enough for 100 ligations). Incubation at 37 °C for 40 min and heat inactivation at 80 °C for 20 min. Done for backbone with and without lacI. Freezed at -20 °C.
Restriction with Spe and EcoRI of each promotor in 1 preparation with 20 µl volume each (enough for 10 ligation for each promotor). Incubate at 37 °C for 40 min and heat inactivation at 80 °C for 20 min. Freezed at -20 °C.
For [http://igem.rwth-aachen.de/2015/index.php/Manual/LigationBB-Assembly ligation], 2 µl of each DNA part from restriction is necessary.
15-05-12
Biotec lab; Attendens: Laura G., Tobi, Sven
consumable material:
- make ampicillin petri dishes for transformations next week
Promotor library:
Dilution of J23113 and -16
Digestion of Promotors 00-19 and heat inactivation (restriction with Spe and EcoRI of each promotor in 1 preparation with 20 µl volume each (enough for 10 ligation for each promotor). Incubation at 37 °C for 40 min and heatinactivate at 80 °C for 20 min).
for [http://igem.rwth-aachen.de/2015/index.php/Manual/LigationBB-Assembly ligation] is 2 µl of each DNA part from restriction necessary. Every restricted promotor is ligated in new backbone with lacI. Everything is freezed -20 °C in Falkon #2.
next steps
finish ligation of J23108, J23109, J23111, J23115 and J23119 in original J61002.
transformation of ligation products in competent DH5α cells (on monday)
15-05-13
Biotec lab; Attendens: Laura G., Tobi, Sven
consumable material:
Promotor library:
Ligation of J23108,-09,-11,-15,-19 in the originial backbone J61002 (without LacI), Incubation for 45 minutes
Heat inactivation for 20 minutes
next steps
15-05-18
Biotec lab; Attendens: Laura G., Tobi
consumable material:
DH5α
Promotor library:
Heatshock transformation of ligation products in DH5α , plate them on Amp-plates (clones with new lacI promotor should be colorless, the five clones with original promotors should be as red as their strength). We used 3 µl of ligation product on ca. 50 µl of competent cells. After 30 minutes ice, 45 seconds water bath, 2 minutes ice we added 200 µl of preheated SOC medium. Incubation for 1 hour (37 °C /250 rpm)
Plates incubate at 37 °C since 16:50.
next steps:
pick single clones from all transformations (19+4) for overnight cultures.
15-05-19
Biotec lab; Attendens: Laura G., Tobi, Sven
consumable material:
new LB-Amp-plates
Promotor library:
In generel poor cell growth but there are colonies on every plate. However, also the 5 promotors (08,09,11,15,19) in the original backbone (without LacI binding site) are white. They should be red. They might turn to red with more time?
We did masterplates (with 6 clones of each) of all transformations. J23102 and -09 only had as few colonies as there are spread on the masterplates.
We make overnight cultures from J23108 ori, -15 ori, -19 ori, -11 ori, -06, -17, -16, -05. "Ori" stands for the original J61002 backbone without LacI binding site.
Next steps:
Do cryos, plasmid preps and prepare sequencing from the overnight cultures (results might come on Friday)
Colony PCR from masterplates (on Thursday):Which primers would we use to check the orientation of our insert?
15-05-20
Biotec lab; Attendens: Laura G., Tobi
consumable material:
Promotor library:
Masterplates:
| promoter |
strength |
Clone colours |
considerations
|
| J23100 |
2547 |
all red |
|
| J23101 |
1791 |
1-4 red |
|
| J23102 |
2197 |
4 red |
|
| J23103 |
17 |
all white |
because of LacI or weak promotor?
|
| J23104 |
1831 |
3-5 red |
|
| J23105 |
623 |
2 red |
|
| J23106 |
1185 |
2,4 red |
|
| J23107 |
908 |
4,5 red |
|
| J23108 |
1303 |
3,6 red |
|
| J23109 |
106 |
only 2 clones, both white |
|
| J23110 |
844 |
1 red |
|
| J23111 |
1487 |
1 red |
|
| J23113 |
21 |
2,5,6 red |
|
| J23114 |
256 |
all white |
|
| J23115 |
387 |
3 red |
|
| J23116 |
396 |
all white |
|
| J23117 |
162 |
all white |
|
| J23118 |
1429 |
2,3 red |
|
| J23119 |
strongest |
all white |
|
| J23119 ori |
strongest |
all red |
|
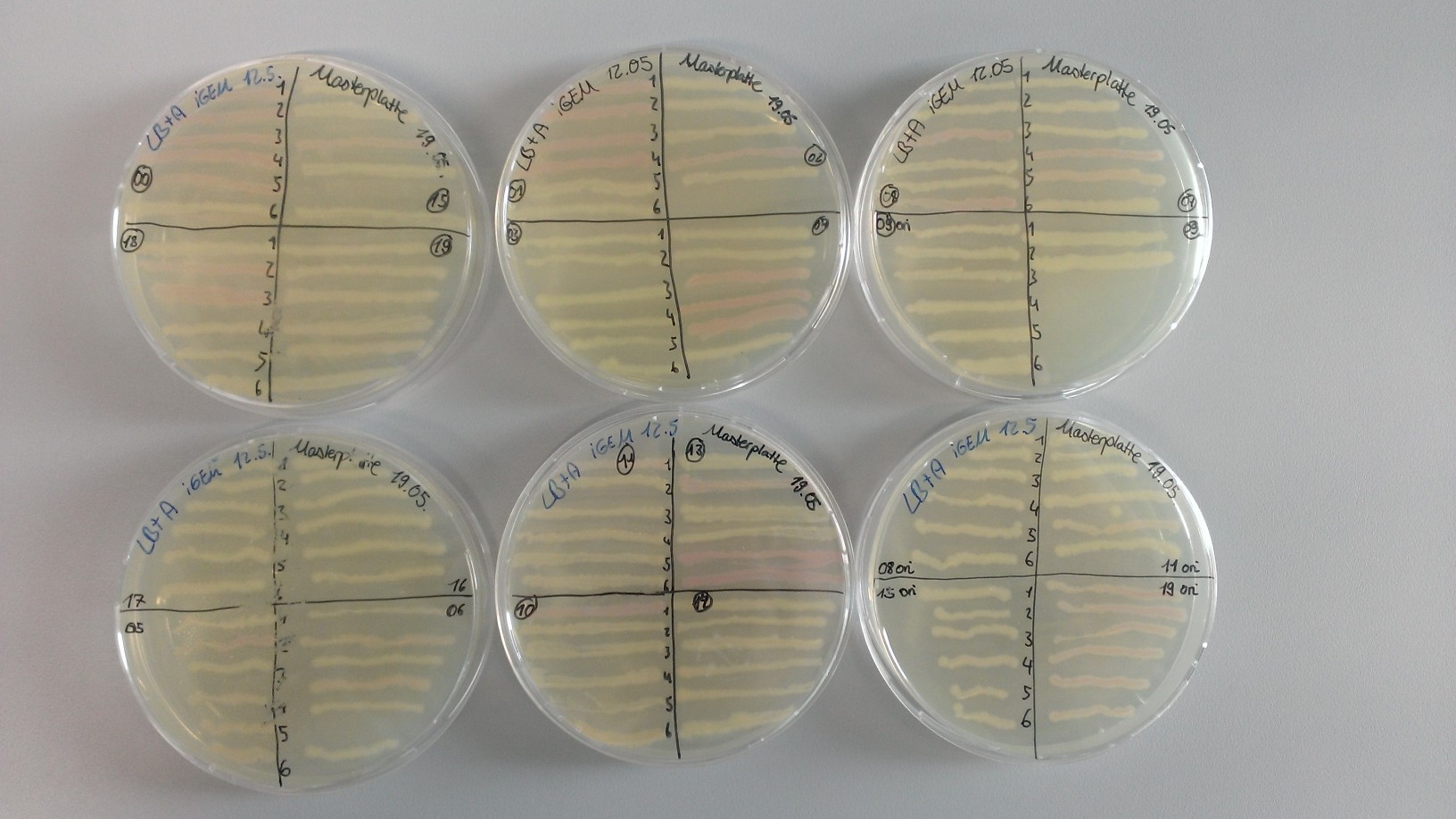
Cryo and plasmidprep from 2 clones (each) of 08 ori, 15 ori, 19 ori, 11 ori, 06, 17, 16, 05.
for sequencing: 05#2, 11ori#2, 16#2, 17#2, 19ori#2 (#4FYS#, #KFWP#, #1CSA#, #ZXCE#, #9TVM#)
15-05-21
Biotec lab; Attendens: Laura G., Tobi, Sven
consumable material:
Promotor library:
DO overnights from all missing constructs (2 clones each) and do cryo of all tomorrow. Do some plasmid preps and freeze the rest of the cultures for plasmid preps next tuesday.
Primer 39OW was designed for sequencing from RFP reverse (necessary for vector library).
15-05-22
Biotec lab; Attendens: Laura G., Tobi
consumable material:
Promotor library:
Make cryo of all overnights.
16 clones are freezed in eppis (-20 °C) for plasmid prep on tuesday. Clones: 13#4, 19#1, 03#2, 09ori#4, 19#2, 14#2, 13#3, 10#2, 01#4, 15#4, 09#1, 14#1, 18#5, 01#5, 09ori#5, 11#6.
16 clones are prepped today and the plasmids are freezed. clones: 08#4, 04#2, 07#2, 09#2, 07#3, 11#5, 15#6, 02#2, 10#3, 00#4, 02#1, 18#4, 00#6, 08#5, 03#1, 04#1.
next steps: on Tuesday
plasmid prep of the last 16 clones.
check sequencing results of the 5 first promotorconstucts.
on Wednesday: prepare 43 plasmids for sequencing.
15-05-26
Biotec lab; Attendens: Sven, Tobi
consumable material:
Promotor library:
16 clones plasmidprep. clones: 13#4, 19#1, 03#2, 09ori#4, 19#2, 14#2, 13#3, 10#2, 01#4, 15#4, 09#1, 14#1, 18#5, 01#5, 09ori#5, 11#6.
check sequencing results of the 5 first promoter constucts.
- #DHAY# expected: J23111 (original), result: between Prefix and Promotor are 18 bp too much (including an unwanted SpeI), between Promotor and RBS are 11 bp too much (including SpeI).
- #HMZF# expected: J23119 (original), result: sequence of J23102 original
- #9TVM# expected: K1585105 (05+lacI), result: sequence of J23102 original
- #1CSA# expected: K1585116 (16+lacI), result: successful!!!
- #MELT# expected: K1585117 (17+lacI), result: nonsense chromatogramme
next steps:
on Wednesday: prepare 43 plasmids (rest of AP+lacI) for sequencing. (see Inventory)
15-05-27
Biotec lab; Attendens: Sven, Tobi
consumable material:
Promotor library:
prepare 43 plasmids (rest of AP+lacI) for sequencing.
next steps
15-05-28
Biotec lab; Attendens: Sven, Tobi, Laura
consumable material:
Promotor library:
try to understand sequencing result of #DHAY#
next steps:
15-05-29
Biotec lab; Attendens: Sven, Tobi, Laura
consumable material:
Promotor library:
We checked the sequencing results. 16 of 24 promoters are successfully built.
next steps:
discard unnecessary tubes with plasmids and cryos after sequencing results.
15-06-01
Biotec lab; Attendens: Sven, Tobi, Laura
consumable material: LB + Amp plates, LB
Promotor library:
new transformation of stored ligation products: J23108 (original) J23109 (original) and BBa_K1585109. Plates incubate since 14:30.
Ligated constructs in [http://tubefront.mosthege.net/iGEM_Aachen/editbox.php?id=FOLKON%20NR.2 Falcon #2].
new overnights from masterplates: 07 (#1, #6), 08 (#1, #2), 11 (#2, #3, #4), 14 (#4, #5, #6), 11ori (#5) (for sequencing) incubate since 15:30.
Discard from Box C 7 tubes, Box D 17 tubes, Box A 14 tubes and Box E 10 tubes.
next steps:
cryos and plasmid prep from overnights for sequencing.
overnights from transformations
15-06-02
Biotec lab; Attendens: Sven, Tobi, Laura
consumable material:
Promotor library:
Cryos (in Box B) and plasmid prep from 07 #1 #6, 08 #1 #2, 11 #2 #3 #4, 14 #4 #5 #6, 11 ori #5
Masterplates and overnights from 08 ori, 09 ori and 09 transformations. Incubation since 14:40.
15-06-03
Biotec lab; Attendantss: Sven, Tobi, Laura
consumable material:
Promotor library:
Cryos (in Box B) and plasmid prep from 08 ori #1, 09 ori #1 (since these were red on the masterplate) and 09 (#1-3)
Sequencing of the above
| Promotor+Clone |
Plasmid |
Cryo
|
| 09 #1 |
#MXEY# |
#OW1V#
|
| 09 #2 |
#FWRP# |
#BS8O#
|
| 09 #3 |
#NWV8# |
#4AEQ#
|
| 09ori #1 |
#ZWAX# |
#TYLE#
|
| 08ori #1 |
#FM8N# |
#BQKN#
|
15-06-11
Biotec lab; Attendants: Sven, Laura
consumable material:
Promotor library:
Sequencing results were aligned to planned sequences. All samples were negative. K1585114 und K1585107 had a point mutation in every promoter. We assume this mutation was introduced by PCR since it is present in all otherwise positive samples of the respective construct. A detailed overview can be found under sequencing results.
15-06-15
Biotec lab; Attendants: Sven
consumable material: new LB + A agar plates, new dilutions of A9W9 and XE3D (10µM, can be found in P' as usual)
Promotor library:
We did another PCR to amplify J23107 (#MAMH#)and J23114 (#6ORC#)to hopefully get rid of the mutation.
DpnI digestion and purification: product can be found at #DMLM# and #O34K#.
Restriction and ligation of J23108, 09, 11 and K1585107, 08, 09, 11, 14 were repeated. Digestion was repeated for all inserts. The old already cut backbones were used (#9P6B# and #FD1K#). Digested inserts can be found in Falcon #FAKE#. Ligated Products can be found in Falcon #FUCK#.
Transformation into E. coli DH5α. Incubation at 37 °C over night.
Plates of K1585119 (#CKYR#) and J23119 (#BYS8#) from cryos for further testing.
15-06-16
Biotec lab; Attendants: Sven
consumable material: IPTG (50x) aliquots, can be found in the antibiotics box (bio 6)
Promotor library:
Master plates and overnight cultures of J23108, 09, 11 and K1585107, 08, 09, 11, 14.
Overnight cultures of J23119 and K1585119. K1585119 was cultivated with and without IPTG to test the expression in comparison to the constitutive J23119.
Cultures of J23108 and J23111 contructs looked slighty reddish on the transformation plates. This is expected since they are both medium strength promoters.
15-06-17
Biotec lab; Attendants: Sven
consumable material:
Promotor library:
We discarded cryos and purified plasmids of the last unsuccessful sequencing.
Plasmid preps of J23108, 09, 11 and K1585107, 08, 09, 11, 14. Three clones each.
Prepared sequencing samples of those constructs
Comparison between J23119, K1585119 and K1585119 + IPTG.
The constitutive promoter J23119 showed a strong rfp expression as expected. Both K1585119 cultures showed only a very weak expression. Repression via the LacI binding site therefore seems to be working as intended whereas the induction with IPTG failed. This may be caused by a too low IPTG concentration. To ensure this the experiment has to be repeated with multiple IPTG concentrations.
- Note: It took quite a long time till rfp expression was visible, even the J23119 culture was still not red after 16h incubation.
15-06-18
Biotec lab; Attendants: Sven
consumable material:
Promotor library:
To Do:
Seperate sequencing table for discarded constructs
new overnight cultures of J23119 and K1585119 with varying IPTG concentrations (0.1; 0.5; 1.0; 2.5; 5 and 10 mM)
15-06-21
Biotec lab; Attendants: Sven, Laura
consumable material:
Promotor library:
Sequencing results were all negative
IPTG induced cultures did not show any red color for all tested IPTG concentrations. We had to do a reverse sequencing to verify that rfp coding sequence is intact.
15-06-22
Biotec lab; Attendants: Sven, Laura
consumable material:
Promotor library:
New PCR for J23108 (PCR with W1KW),J23109(PCR with W49P) and J2311 (PCR with S8R6) was not successful.
Gel of restricted promotors showed nonsense (reason for nonsense ligation products)
backbone: restriction of 3N4M (once with Eco/Xba and once with Eco/Spe). Both tubes are in Falcon "22.06. Back"
prepare sequencing of successful promoters with reverse primer (XE3D) since IPTG induction failed
To Do:
Do we have to amplify backbone once more??
15-06-23
Biotec lab; Attendants: Sven, Laura
consumable material:
Promotor library:
We did new PCRs of J23107, 08, 09, 11, 14
However, the gel showed hardly any bands. Because the PCR components seem to be fine, we have to use a new, prepped template.
15-06-24
Biotec lab; Attendants: Sven, Laura
consumable material:
Promotor library:
We did plates of the original kitplate transformants for J23107 (#X4YM#), J23108 (#XXM6#), J23109 (#KS96#), J23111 (#MBKL#) and J23114 (#XBVM#).
note: J23108, J23109 and J23111 in pSB1C3
15-06-25
Biotec lab; Attendants: Sven, Laura
consumable material:
Promotor library:
Preparation for induction tests: plates and overnights of J23119 (#BYS8#) and K1585119 (#CKYR# and #YFV9#).
plates from cryo and overnight cultures of J23107, J23108, J23109, J23111 and J23114.
15-06-26
Biotec lab; Attendants: Tobi, Laura
consumable material:
Promotor library:
Result of induction test: (#YFV9# not ind; #YFV9# ind; #BYS8#; #CKYR# ind; #CKYR# not ind)
Plasmidprep of overnights (#DE6O#, #BRRS#, #CXSS#, #PT93#, #DMCX#)
PCR and gel verification.
15-06-29
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
PCR of #PT93# and #DMCX# with A9W9/XE3D to amplify J23111 and J23114 inserts.
DpnI digestion of J23107, J23108, J23109 (pcr 2015-06-26) and J23111, J23114 (see above).
The gel showed only light bands. The purification of digested PCR products was not successful. Almost no DNA measured, only salt peak at 230 nm.
Repeat full PCR for J23107 (#DE6O#), J23108 (#BRRS#), J23109 (#CXSS#), J23111 (#PT93#) and J23114 (#DMCX#). (see 2015-06-26)
Gel of full repeat of PCR promoter amplification
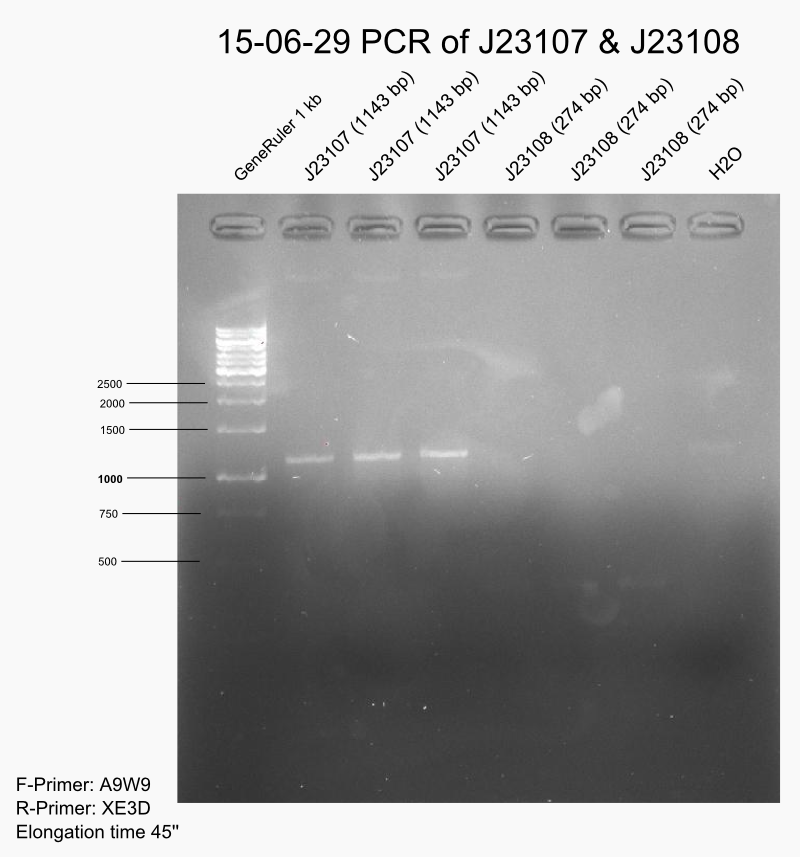
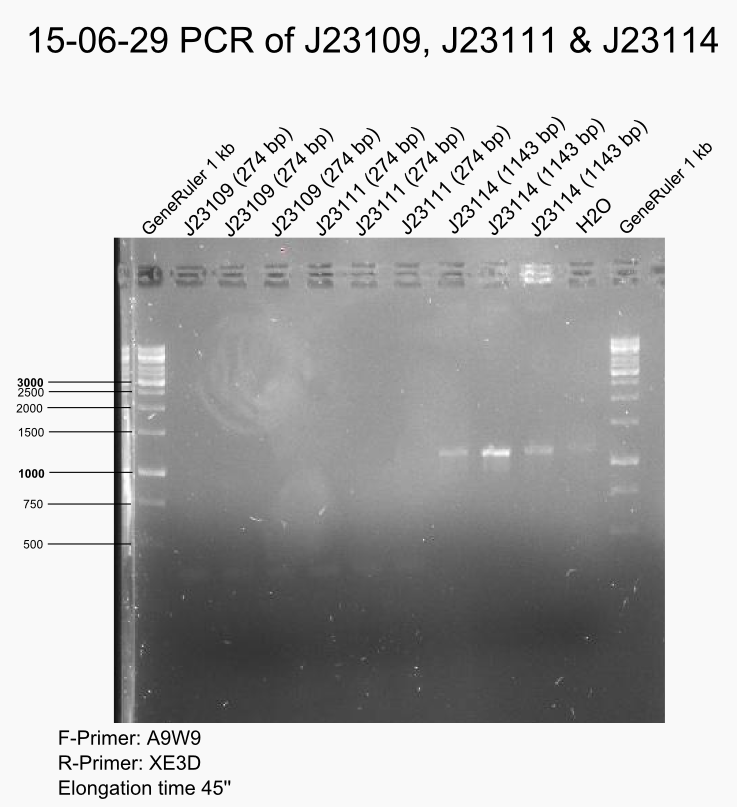
Characterisation
Overnights of J23119 (constitutive, #BYS8#), K1585119 induced and non-induced (#CKYR# and #YFV9#) and pSB1K30 (positive control) to verify whether or not our IPTG stock is fine.
Induced cultures were prepared in 3 concentrations each: 0.2x (1 µM), 1x (5 µM) and 5x (25 µM).
pSB1K30 in NEB Turbo is used as positive control. Note: Plasmid with kanamycin resistance!
Incubation at 37 °C for 18 h.
15-06-30
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
another PCR of all 5 promotors: 30 cycles and 2ul template
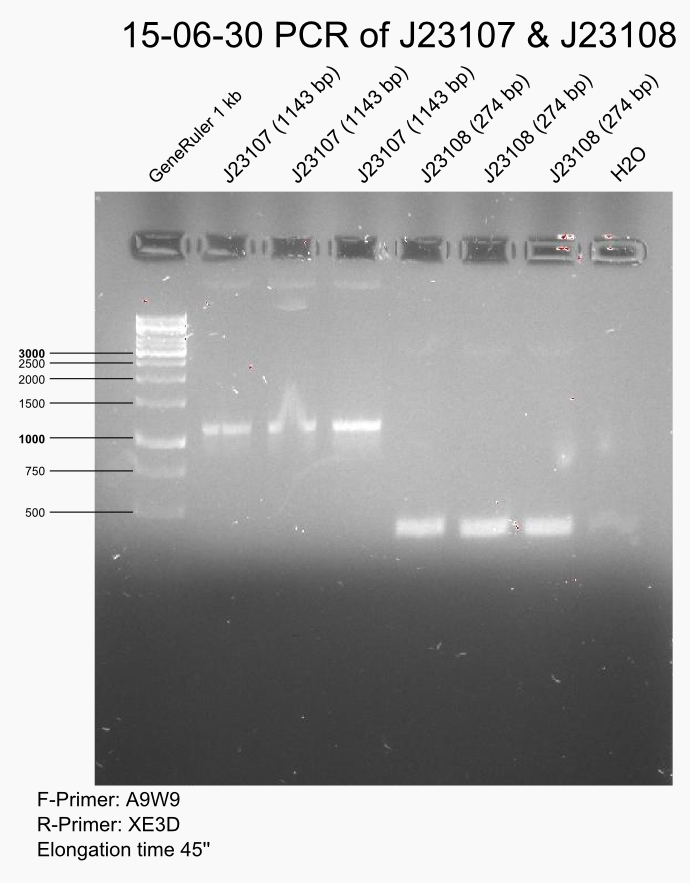
Gel of J23109, -11 & -14 is nonsense.
Do DpnI digest of J23107 & J23108 for 1,5h plus heat inactivation.
Result of induction test (only visual with green light):
constitutive J23119 has the strongest red colour. Not induced K30 & AP19 are colourless. Strain #CKYR# and #YFV9# have always the same colour under the same conditions. Induced K30 is always a little less colourful as the induced AP19s (that is unexpected). 0,2x IPTG shows no difference to not induced cultures. 1x IPTG is slightly red. 5x IPTG have red colour but not as strong as the constitutive one.
Experiment gives hardly any usable information. J23119 should clearly show a strong pink/red color visible in regular light (see documentation 15-06-17). The negative result of the J23119 implies that there might be some problems with cultivation. Cultures might not get enough oxygen to form the rfp (people at bio6 sometimes forget to switch on the shaker after they removed their samples).
'New Induction Test':
Same setup as before. This time we used shakers at the iAMB. Both IPTG stocks are tested at the same time
Overnights of J23119 (constitutive, #BYS8# from 2 different plates), K1585119 induced (3 times) and non-induced from #CKYR# and pSB1K30 in NEB Turbo ## induced 3 times and one not induced (Note: Plasmid with kanamycin resistance!)
For induction we used two different IPTG stocks 50x & 1000x.
Induced cultures were prepared in 3 concentrations each: 1x (5 µM), 5x (25 µM) and 10x (50 µM).
Incubation at 37 °C starting at 16:00.
Results:
J23119 cultures both show a very strong pink color after 24 h of incubation. All other cultures do not show a significant amount of rfp expression. Positive control is negative due to the use of a wrong strain (NEB Turbo is a cloning strain without T7 polymerase)
Next time we use BBa_I13507 in pSB1A30 in BL21 with Cas9 to express RFP (Cas9 will be destroied at 37 °C) (alternatively BBa_I13507 in pSB1A30 in BL21 is in Cryo #6DA3#). And pSB1K30 in BL21 (#KK16#).
15-07-01
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
Purification of J23107 & -08 PCR products is successful (#3X3H#, #Z6MT#).
Restriction of J23107 & -08 with EcoRI and SpeI (stored #WT6F#). Gel control of restriction.

=> ligation with backbone with and without LacI binding site (stored #DWCR#).
Transformation of ligation products (AmpR) 07 & 08 mit LacI backbone, 08 in original backbone.
Induction Test:
Repeat of the previous induction experiment. This time we used pSB1K30 and pSB1A30 (both induced 1x and not induced to test). One J23119 culture was used as positive control for general rfp expression.
Results:
Positive control was negative even after 40 h. Cultivation in bio6 shakers probably does not provide enough oxygen to enable rfp processing. We will try to do a first real characterization experiment in deep well plates next week. Aerobic conditions should not be a problem there.
next steps
Masterplates of transformants.
PCR of J23109, -11 & -14
15-07-02
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
PCR of J23109, -11 & -14. Gel control.
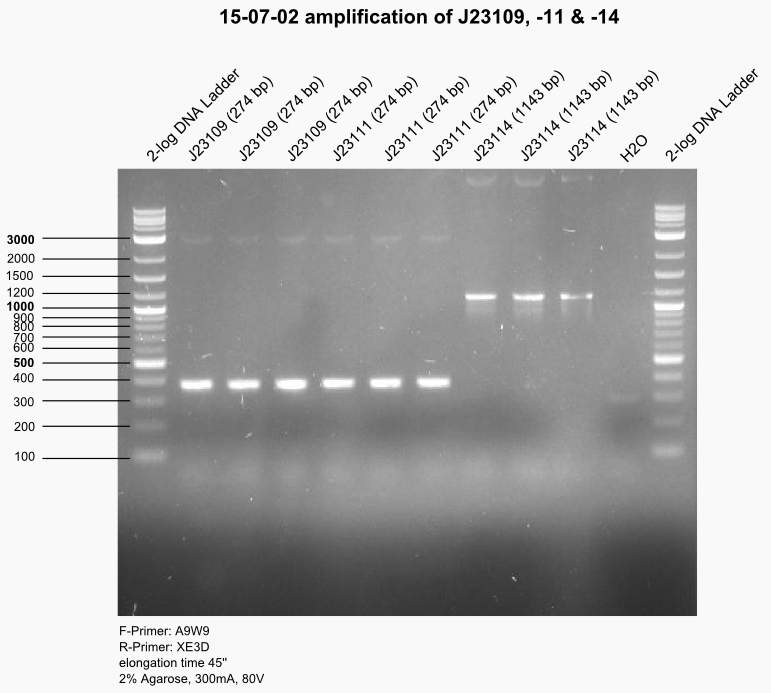
DpnI digest and heat inactivation #Z1AR#, #1AWV#, #ZBWQ#. Digest everything with EcoRI & SpeI and ligate to backbone with and without LacI binding site. Transform all ligation products.
Do overnights and masterplates (pick lots of clones) of successful transformations (only J23108 and K1585107).
15-07-03
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
We made cryos and plasmid prep of overnight cutures (K1575107 #1-8 & J23108 #1-8).
next steps (on monday)
prepare for sequencing:
#84YY#
#3X4N#
#S3Z3#
#EY1D#
#BOBB#
#HQBY#
#HPP3#
#HWKM#
#CZEK#
#KYYR#
#Y9H4#
#Y6FZ#
#VEEK#
#E6F1#
#6N6W#
#1836#
15-07-04
Biotec lab; Attendants: Sven, Laura
consumable material:
Promotor library:
Cryo of overnight cultures and freezing of the cultures for plasmidprep on monday.
15-07-06
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
Plasmidprep of J23109, -11 & K1575109, -14
Prepare sequencing of J23108, J23109, J23111 & K1575107, K1575109, K1575114
15-07-07
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
We plated all constitutive and inducible cryos with J61002 backbone on LB+A.
15-07-08
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
Check last sequencing: no successful construct!!!
For characterization: Do overnights in wellplate of J23100, J01, J02, J03, J04, J05, J06, J10, J13, J15, J16, J17, J18 & J19 and K15751xx versions of each. Incubation started 18:30 and 37 °C.
next steps
Add glycerol to overnights for freezing.
discard plasmid in -20 °C: J23108, J23109, J23111 & K1575107, K1575109, K1575114 from last week.
discard cryos in -80 °C: J23108, J23109, J23111 & K1575107, K1575109, K1575114 from last week.
15-07-09
CHARACTERIZATION
Biotec lab; Attendants: Sven, Tobi, Laura
consumable material:
Promotor library:
We did a cryo wellplate with all verified promoters: 90 µl of 50 % glycerol to the overnight wellplate. Freezed in -80 °C in Bio7.
From this cryo wellplate we inoculated 2 wellplates with LB+Amp, one of these plates with IPTG.
We measured fluorescence and OD in the platereader every hour. We measured both OD and fluoresence to understand a possible correlation between both factors.
Note: mRFP has its excitation peak at 584nm and its emission peak at 607nm wavelength.
15-07-14
Biotec lab; Attendants: Tobi, Laura
consumable material:
Promotor library:
Characterization:
We added 200 µl LB media per well in grey SystemDuetz plates. Ampicilin was added in each well except from 6 C-H & 12 C-H. In one wellplate IPTG (20 µl/20 ml x1000stock). Inoculation via cyroreplicator. Add 3 µl DH5α (as wildtype control) from overnight in 6C+D & 12C+D.
We did not do a measurement at the beginning because grey plates do not fit in the platereader. For measurement the cultures get transfered into black wells with glas bottom.
Incubation at 30 °C and 300 rpm started at 10:10 a.m.
We did an OD/F measurement at 19:00 because there should be some spots with only LB media as reference.
15-07-15
Biotec lab; Attendants: Tobi, Laura
consumable material:
Promotor library:
Characterization:
We did another OD/F measurement again in the morning at 8:15.
Sterilisation of 2 grey and 2 black wellplates with glassbottoms.
15-07-17
OD/F measurment was started. Measurement every 2-3 hours.
15-07-18
Inoculation of one plate in the morning. Measurements every hour.
15-07-19
Last measurement was done in the morning.
15-07-20
Do overnights of J23100, J01, J02, J03, J04, J05, J06, J10, J13, J15, J16, J17, J18 & J19 and corresponding K15751xx versions of each. Incubation started at xxh in iAMB and 37 °C.
15-07-21
We did a new Cryo wellplate with new layout.
15-07-22
New measurement plates. Incubation at 30 °C and 300 rpm (150 ul in each well) since 14 o'clock.
15-07-23
First measurement at 9:00. Induction of IPTG wellplate. Shaking started at 9:40.
Measure each hour. At 10.45; 12.00; 13.15; 14.30; 15.45; 17.00 & 18.15
Result
The characterized promoters are:
J23100, J23101, J23102, J23103, J23104, J23105, J23106, J23110, J23113, J23115, J23116, J23117, J23118 & J23119
The native Anderson Promoters show the expected relative fluorescence.
Plotting of the files:
The characterized promoters with lacI binding site are:
K1575100, K1575101, K1575102, K1575103, K1575104, K1575105, K1575106, K1575110, K1575113, K1575115, K1575116, K1575117, K1575118 & K1575119.
Results Summary
14 different Anderson Promoters are present in backbone J61002 with RFP (E1010) as control between the SpeI & PstI restriction sites.
These 14 Anderson Promoters are also present with an lacI-binding site.
The inhibition by lacI works perfectly.
The induction with IPTG is not significantly possible.