Difference between revisions of "Team:Exeter/Results"
| (3 intermediate revisions by the same user not shown) | |||
| Line 86: | Line 86: | ||
captions: true, | captions: true, | ||
adaptiveHeight: true, | adaptiveHeight: true, | ||
| − | preloadImages: 'all' | + | preloadImages: 'all', |
| + | pager: 'false' | ||
}); | }); | ||
| Line 96: | Line 97: | ||
captions: true, | captions: true, | ||
adaptiveHeight: true, | adaptiveHeight: true, | ||
| − | preloadImages: 'all' | + | preloadImages: 'all', |
| + | pager: 'false' | ||
}); | }); | ||
| Line 106: | Line 108: | ||
captions: true, | captions: true, | ||
adaptiveHeight: true, | adaptiveHeight: true, | ||
| − | preloadImages: 'all' | + | preloadImages: 'all', |
| + | pager: 'false' | ||
}); | }); | ||
| Line 168: | Line 171: | ||
<h4><em>In silico</em> testing:</h4> | <h4><em>In silico</em> testing:</h4> | ||
| + | |||
| + | |||
| + | To see an explanation of <em>in silico</em> testing, please visit the <a href="https://2015.igem.org/Team:Exeter/Modeling">modelling page</a>. | ||
| + | |||
| + | Download raw data files for Zeus NUPACK testing <a href="https://static.igem.org/mediawiki/2015/c/c3/Exeter_ZEUSJ_NuPACK.zip">here</a>. | ||
| + | Visit the <a href="http://parts.igem.org/Part:BBa_K1586003">registry page</a> for more information. | ||
</div> | </div> | ||
| Line 220: | Line 229: | ||
<div id="further_characterisation"> | <div id="further_characterisation"> | ||
| + | <div style="float:right"> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/b/bb/Exeter_chromoplates.png" width="200"> | ||
| + | |||
| + | </div> | ||
<h2>Further characterisation:</h2> | <h2>Further characterisation:</h2> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<p> | <p> | ||
| Line 258: | Line 266: | ||
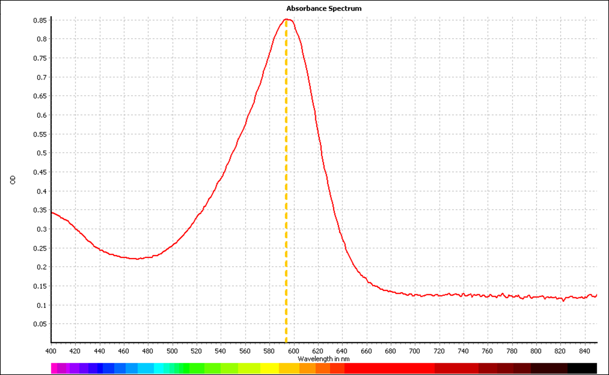
Shown in figure 4 are the absorbance spectra for aeBlue, eforRed, and amajLime between 400nm and 850nm. The peak maxima for these are 595nm, 580nm, and 595nm respectively. | Shown in figure 4 are the absorbance spectra for aeBlue, eforRed, and amajLime between 400nm and 850nm. The peak maxima for these are 595nm, 580nm, and 595nm respectively. | ||
</p> | </p> | ||
| + | </br> | ||
| + | </br> | ||
</br> | </br> | ||
</br> | </br> | ||
| Line 286: | Line 296: | ||
</br> | </br> | ||
| − | <h4> | + | <h4>Visual limits:</h4> |
| + | |||
| + | <div class="containerSlider300" style="float:left"> | ||
| + | <div class="slider300"> | ||
| + | |||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/4/4a/Exeter_amajLime_visual_limit.png"><img src="https://static.igem.org/mediawiki/2015/4/4a/Exeter_amajLime_visual_limit.png" title="Figure 6: Visual limits for amajLime."></a></div> | ||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/1/18/Exeter_aeBlue_visual_limit.png"><img src="https://static.igem.org/mediawiki/2015/1/18/Exeter_aeBlue_visual_limit.png" title="Figure 6: Visual limits for aeBlue."></a></div> | ||
| + | <div class="slide"><a href="https://static.igem.org/mediawiki/2015/2/23/Exeter_eforRed_visual_limit.png"><img src="https://static.igem.org/mediawiki/2015/2/23/Exeter_eforRed_visual_limit.png" title="Figure 6: Visual limits for eforRed."></a></div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <p> | ||
| + | Figure 6 shows qualitative data for the colour intensities of the chromoproteins at 50 μg, 10 μg, 5 μg, and 1 μg in 50 μl volumes. | ||
| + | </p> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
| + | |||
| + | |||
</div> | </div> | ||
Latest revision as of 16:55, 1 October 2015
Results
Toehold experiments
In silico testing:
To see an explanation of in silico testing, please visit the modelling page. Download raw data files for Zeus NUPACK testing here. Visit the registry page for more information.Experimental validation of GreenFET1J (BBa_K1586000):
As was explained on the experiments page, GreenJ's (BBa_K1586000) function was characterised and validated by measuring fluorescence intensity of GFP in response to increasing amounts of trigger RNA. Shown in figure 1 is the raw data which was collected from this experiment. In order to analyse this data, the log10 of the amounts of TrigGreen (in nanograms) were calculated. So that the log10 value for 0ng could be calculated, 1x10-6ng was used instead, and all values were increased by six so that 0ng shows as 0 on the graph. These data were then plotted on a graph and a linear trend-line fitted (figure 2).

Further characterisation:
The following parts were further characterised:
Absorbance spectra:
Shown in figure 4 are the absorbance spectra for aeBlue, eforRed, and amajLime between 400nm and 850nm. The peak maxima for these are 595nm, 580nm, and 595nm respectively.
Standard curve - concentration vs. OD:
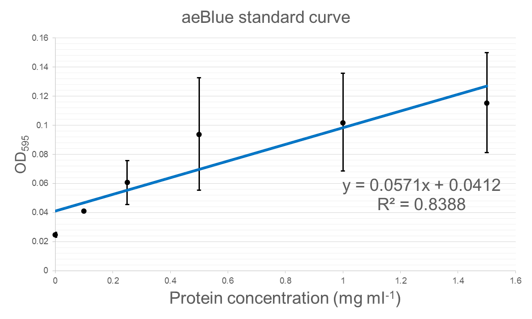
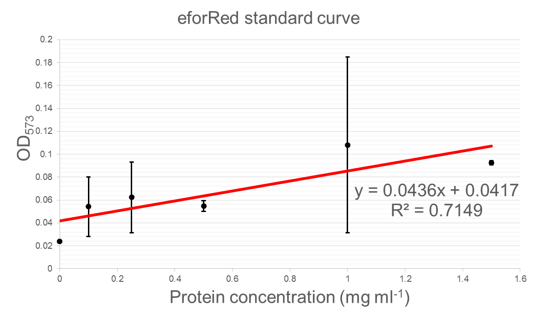
Figure 5 shows standard curves of concentration in mg ml-1 vs. OD at peak maxima for each of the three chromoproteins further characterised.
Visual limits:
Figure 6 shows qualitative data for the colour intensities of the chromoproteins at 50 μg, 10 μg, 5 μg, and 1 μg in 50 μl volumes.