Difference between revisions of "Team:Heidelberg/Overview"
| Line 10: | Line 10: | ||
</div> | </div> | ||
</body> | </body> | ||
| + | </html> | ||
| + | <html> | ||
| + | <head> | ||
| + | <script src="jquery.js" type="text/javascript" charset="utf-8"></script> | ||
| + | <script src="raphael.js" type="text/javascript" charset="utf-8"></script> | ||
| + | |||
| + | <script type="text/javascript"> | ||
| + | var flagCanvasPath = 'M7.753,64.331c-0.02-0.055-0.031-0.314-0.139-0.361c-0.746-0.338-1.551-0.945-2.258-0.852c-0.686,0.092-1.014-0.26-1.469-0.449c-0.514-0.215-1.035-0.506-1.424-0.893c-0.219-0.219-0.229-0.695-0.246-1.061c-0.084-1.984-0.131-3.969-0.207-5.953c-0.025-0.609-0.127-1.217-0.156-1.828c-0.064-1.41-0.104-2.818-0.162-4.229c-0.098-2.459-0.217-4.916-0.291-7.375c-0.008-0.299,0.217-0.602,0.322-0.906c0.049-0.139,0.109-0.289,0.1-0.428c-0.086-1.037-0.189-2.072-0.281-3.109c-0.209-2.309-0.439-4.615-0.604-6.924c-0.117-1.652-0.131-3.309-0.219-4.961c-0.057-1.063-0.191-2.121-0.24-3.182c-0.045-0.924-0.01-1.852-0.023-2.777c-0.047-2.795-0.08-5.592-0.164-8.387C0.237,8.841,0.104,7.024-0.001,5.118c1.063-0.256,2.156-0.188,3.158,0.119c1.34,0.41,2.605,1.061,3.914,1.58c1.83,0.725,3.645,1.502,5.672,1.516c1.152,0.006,2.305,0.09,3.455,0.146c2.031,0.102,3.828-0.59,5.578-1.549c1.484-0.813,3.082-1.336,4.783-1.566c1.4-0.191,2.773-0.551,4.17-0.779c1.166-0.189,2.346-0.318,3.523-0.426c0.225-0.021,0.469,0.174,0.756,0.291c0.117-0.141,0.25-0.303,0.324-0.393c0.291,0.098,0.502,0.227,0.713,0.229c1.145,0.021,2.291-0.057,3.43,0.023c2.145,0.152,4.273,0.424,6.334,1.117c2.813,0.947,5.635,1.879,8.475,2.752c0.74,0.227,1.555,0.248,2.34,0.291c1.564,0.08,3.129,0.154,4.695,0.154c3.439,0,6.756-0.711,9.859-2.176c2.021-0.951,3.918-2.164,5.885-3.236c0.99-0.541,2.002-1.045,3.021-1.531c0.234-0.111,0.557-0.059,0.385-0.471c0.238-0.053,0.471-0.123,0.711-0.152c0.285-0.037,0.572-0.041,0.861-0.057c-0.004-0.068-0.008-0.137-0.014-0.203c-0.123,0-0.246,0-0.369,0c-0.014-0.043-0.023-0.086-0.037-0.129c0.801-0.186,1.602-0.357,2.396-0.559c1.615-0.402,2.84,0.35,3.965,1.348c0.211,0.186,0.314,0.492,0.455,0.748c0.168,0.295,0.027,0.674,0.51,0.914c0.271,0.137,0.326,0.889,0.318,1.361c-0.014,1.105-0.381,2.279-0.119,3.299c0.311,1.219-0.365,2.074-0.574,3.086c-0.221,1.082-0.629,2.121-0.912,3.191c-0.736,2.824-1.48,5.648-2.16,8.486c-0.369,1.543-0.645,3.111-0.91,4.676c-0.09,0.557,0.008,1.143-0.029,1.711c-0.195,2.98-0.336,5.967-0.637,8.936c-0.266,2.598-0.076,5.158,0.348,7.699c0.373,2.236,0.832,4.469,1.449,6.646c0.332,1.174,1.162,2.199,1.592,3.355c0.369,0.996,0.563,2.074,0.688,3.133c0.197,1.641-0.453,3.141-1.061,4.611c-0.244,0.592-0.707,1.104-1.125,1.607c-1.482,1.795-3.367,3.025-5.576,3.703c-0.885,0.271-1.846,0.289-2.771,0.422c-1.982,0.285-3.951,0.258-5.906-0.225c-0.66-0.164-1.344-0.229-2.002-0.398c-1.502-0.393-3.016-0.756-4.486-1.246c-2.246-0.748-4.455-1.604-6.688-2.391c-1.633-0.574-3.254-1.176-4.916-1.654c-1.666-0.477-3.4-0.73-5.059-1.23c-2.094-0.635-4.246-0.445-6.361-0.543c-2.463-0.117-4.953,0.33-7.42,0.225c-2.297-0.1-4.113,1.145-6.174,1.676c-1.805,0.463-3.623,0.85-5.428,1.307c-0.914,0.23-1.791,0.621-2.711,0.805c-1.928,0.381-3.871,0.605-5.84,0.244c-1.885-0.348-3.781-0.656-5.67-0.994C8.485,64.562,8.171,64.448,7.753,64.331z'; | ||
| + | |||
| + | // Used to add a hover action handler for the flag canvas which not only | ||
| + | // hovers the flag canvas but the flag pole as well. | ||
| + | // The normal hover cannot be used on the set flag, as every element of | ||
| + | // the set would get it's own hover handler... and therefore only make | ||
| + | // itself glow. | ||
| + | // This does not register a hover handler for the flag pole and assumes | ||
| + | // flagCanvas == flag[0], flagPole == flag[1], and flagIcon == flag[3]! | ||
| + | function addHover(flag, glowColor) { | ||
| + | var glowColor = glowColor || '#C17D11'; | ||
| + | var flagCanvas = flag[0]; | ||
| + | var flagPole = flag[1]; | ||
| + | var flagIcon = flag[2]; | ||
| + | mouseover = function() { | ||
| + | flagCanvas.g = flagCanvas.glow({color: glowColor, width: 10}); | ||
| + | flagPole.g = flagPole.glow({color: glowColor, width: 10}); | ||
| + | }; | ||
| + | mouseout = function() { | ||
| + | flagCanvas.g.remove(); | ||
| + | flagPole.g.remove(); | ||
| + | }; | ||
| + | |||
| + | flagCanvas.hover(mouseover, mouseout); | ||
| + | flagIcon.hover(mouseover, mouseout); | ||
| + | } | ||
| + | |||
| + | // Call upon document ready | ||
| + | $(document).ready(function() { | ||
| + | // Initialice the map canvas | ||
| + | var map = Raphael('map', 1200, 800); | ||
| + | |||
| + | // Draw the map | ||
| + | var background = map.image('https://static.igem.org/mediawiki/2015/a/af/Heidelberg_map_background.svg', 0, 0, 922, 576); | ||
| + | var ship = map.image('https://static.igem.org/mediawiki/2015/1/10/Heidelberg_map_ship.svg', 60, 80, 115, 95).attr({href: 'https://2015.igem.org/Team:Heidelberg/Team', title: "Team"}); | ||
| + | var star = map.image('https://static.igem.org/mediawiki/2015/5/5e/Heidelberg_map_star.svg', 710, 40, 145, 149); | ||
| + | var island = map.image('https://static.igem.org/mediawiki/2015/9/9a/Heidelberg_map_island.svg', 190, 70, 585, 443); | ||
| + | |||
| + | // Flag prototype | ||
| + | var flag = map.set(); | ||
| + | var flagCanvas = map.path(flagCanvasPath); // The canvas | ||
| + | var flagPole = map.path("M0,10L5,120"); // The pole | ||
| + | flag.push(flagCanvas, flagPole); | ||
| + | flag.attr({'stroke-width': '2px', fill: 'white'}); | ||
| + | |||
| + | // RD, CF, AB, RTSMS, SW6 RFC flags | ||
| + | // Flag and icon can not be preassembled and then moved | ||
| + | // as this would reset the scale transformation on the icon | ||
| + | var flagRD = flag.clone(); | ||
| + | var iconRD = map.image('https://static.igem.org/mediawiki/2015/3/3d/Heidelberg_map_rd_icon.png', 0, 0, 171, 192); | ||
| + | iconRD.transform('t400,280s0.23'); | ||
| + | flagRD.transform('t440,340'); | ||
| + | flagRD.push(iconRD); | ||
| + | addHover(flagRD); | ||
| + | flagRD.attr({href: 'https://2015.igem.org/Team:Heidelberg/project/hlpd', title: "Rape Drugs"}); | ||
| + | |||
| + | var flagCF = flag.clone(); | ||
| + | var iconCF = map.image('https://static.igem.org/mediawiki/2015/5/50/Heidelberg_map_cf_icon.png', 0, 0, 171, 192); | ||
| + | iconCF.transform('t263,-27s0.23'); | ||
| + | flagCF.transform('t310,35'); | ||
| + | flagCF.push(iconCF); | ||
| + | addHover(flagCF); | ||
| + | flagCF.attr({href: 'https://2015.igem.org/Team:Heidelberg/project/cf', title: "Cystic Fibrosis"}); | ||
| + | |||
| + | var flagAB = flag.clone(); | ||
| + | var iconAB = map.image('https://static.igem.org/mediawiki/2015/d/df/Heidelberg_map_ab_icon.png', 0, 0, 171, 192); | ||
| + | iconAB.transform('t458,22.25s0.23'); | ||
| + | flagAB.transform('t500,85'); | ||
| + | flagAB.push(iconAB); | ||
| + | addHover(flagAB); | ||
| + | flagAB.attr({href: 'https://2015.igem.org/Team:Heidelberg/project/ab', Dtitle: "Aptabodies"}); | ||
| + | |||
| + | var flagRTSMS = flag.clone(); | ||
| + | var iconRTSMS = map.image('https://static.igem.org/mediawiki/2015/d/d9/Heidelberg_map_rtsms_icon.png', 0, 0, 220, 181); | ||
| + | iconRTSMS.transform('t580,163s0.23'); | ||
| + | flagRTSMS.transform('t650,220'); | ||
| + | flagRTSMS.push(iconRTSMS); | ||
| + | addHover(flagRTSMS); | ||
| + | flagRTSMS.attr({href: 'https://2015.igem.org/Team:Heidelberg/project/rtsms', title: "Real Time Small Molecule Sensing"}); | ||
| + | |||
| + | var flagSW = flag.clone(); | ||
| + | var iconSW = map.image('https://static.igem.org/mediawiki/2015/6/68/Heidelberg_map_sw_icon.png', 0, 0, 171, 192); | ||
| + | iconSW.transform('t320,100s0.23'); | ||
| + | flagSW.transform('t360,160'); | ||
| + | flagSW.push(iconSW); | ||
| + | addHover(flagSW); | ||
| + | flagSW.attr({href: 'https://2015.igem.org/Team:Heidelberg/Software', title: "Software"}); | ||
| + | |||
| + | var flagRFC = flag.clone(); | ||
| + | var iconRFC = map.image('https://static.igem.org/mediawiki/2015/6/6d/Heidelberg_map_rfc_icon.png', 0, 0, 254, 135); | ||
| + | iconRFC.transform('t180,300s0.23'); | ||
| + | flagRFC.transform('t260,330'); | ||
| + | flagRFC.push(iconRFC); | ||
| + | addHover(flagRFC); | ||
| + | flagRFC.attr({href: 'https://2015.igem.org/Team:Heidelberg/project/standardization', title: "Request For Comment"}); | ||
| + | |||
| + | // Make the prototype flag invisible | ||
| + | flag.attr({opacity: 0}); | ||
| + | |||
| + | // Animate drawing of ship's path | ||
| + | var paths = { | ||
| + | ship2sw: map.path('M148,167c0,0,96.26,4.977,121,17c28.76,13.977,93,87,93,87'), | ||
| + | ship2cf: map.path('M151.333,164.667c0,0,60.348-30.175,79.667-32c21.265-2.008,82.667,16.667,82.667,16.667'), | ||
| + | sw2rtsms: map.path('M367.5,275.5c0,0,124.399-7.397,159,0c36.149,7.729,124,60.5,124,60.5'), | ||
| + | sw2ab: map.path('M367.5,272.212c0,0,67.955-16.567,86.25-25.962c15.643-8.033,49.5-43.75,49.5-43.75'), | ||
| + | sw2rfc: map.path('M364.5,278c0,0,13.354,55.277,10.5,74.5c-2.702,18.197-20.642,52.9-34.5,65c-14.448,12.615-72,26.5-72,26.5'), | ||
| + | sw2rd: map.path('M367.5,278c0,0,17.831,82.886,25.5,101.333C403.623,404.886,440.667,454,440.667,454') | ||
| + | }; | ||
| + | for (i in paths) { | ||
| + | path = paths[i]; | ||
| + | path.attr({opacity: 0}); | ||
| + | } | ||
| + | |||
| + | loaded = [false, false, false, false, false, false, false, false, false, false]; | ||
| + | images = [ship, star, island, background, iconRD, iconCF, iconSW, iconRFC, iconRTSMS, iconAB]; | ||
| + | for (i in images) { | ||
| + | image = images[i]; | ||
| + | image.node.setAttribute('loadId', i); | ||
| + | image.node.setAttribute('externalResourcesRequired','true'); | ||
| + | image.node.addEventListener("load", function() { | ||
| + | loaded[this.getAttribute('loadId')] = true; | ||
| + | }) | ||
| + | } | ||
| + | |||
| + | animateShipPath = function(shipPath, segmentStart, secondLevel) { | ||
| + | if (segmentStart < shipPath.getTotalLength()) { | ||
| + | var subpath = map.path(shipPath.getSubpath(segmentStart, segmentStart + 5)); | ||
| + | subpath.attr({"stroke-width": 2}); | ||
| + | setTimeout(animateShipPath, 50, shipPath, segmentStart + 10, secondLevel); | ||
| + | } else if (secondLevel) { | ||
| + | animateShipPath(paths.sw2rfc, 0, false); | ||
| + | animateShipPath(paths.sw2ab, 0, false); | ||
| + | animateShipPath(paths.sw2rd, 0, false); | ||
| + | animateShipPath(paths.sw2rtsms, 0, false); | ||
| + | } else { | ||
| + | return; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | waitForImages = function() { | ||
| + | var ready = true; | ||
| + | for (i in loaded) { | ||
| + | if (!loaded[i]) { | ||
| + | ready = false; | ||
| + | break; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | if (ready) { | ||
| + | animateShipPath(paths.ship2sw, 0, true); | ||
| + | animateShipPath(paths.ship2cf, 0, false); | ||
| + | } else { | ||
| + | setTimeout(waitForImages, 100); | ||
| + | } | ||
| + | }; | ||
| + | |||
| + | waitForImages(); | ||
| + | }); | ||
| + | </script> | ||
| + | </head> | ||
| + | |||
| + | <body> | ||
| + | <div id="map"></div> | ||
| + | </body> | ||
</html> | </html> | ||
{{Heidelberg/pages/overview/intro}} | {{Heidelberg/pages/overview/intro}} | ||
Revision as of 15:57, 18 September 2015

Diving into the world of functional nucleic acids
In 1952 DNA was described as the carrier of genetic information for the first time.
Catalytic RNA – Ribozymes

The secondary structure of a Hammerhead Ribozyme (HHR). Arrow indicates position of cleavage between third and first stem. First and second stem are stem loops while stem three is open.
In 1982 the first catalytic RNA (Ribozyme): a self-spicing intron from Tetrahymena pre-rRNA was described.
Another ribozyme that is related to the HHR is the hepatitis δ virus ribozyme (HDV).

Naturally occurring Hairpin Ribozyme with cleavage site indicated by arrow.
Both of the mentioned types of ribozymes are found in satellite RNA of plant origin
Our idea of working with functional nucleic acids originated from this system. We were fascinated by the vast variety of processes that they can perform and started digging deeper into the potential of nucleic acids as tools. During this process we came across interesting systems amongst which we found the twin ribozyme (Fig. 3). Another famous hairpin-ribozyme-derived functional nucleic acid developed by Müller.
Next to the directed evolution of existing ribozymes in vitro selection methods

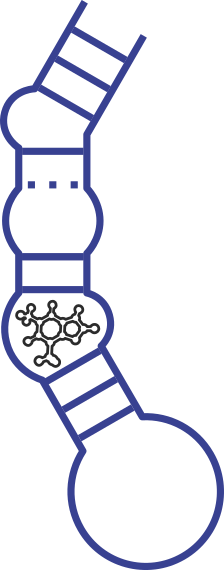
Secondary structure of the Twin Ribozyme. With cleavage and ligation sites indicated by arrows. The Hairpin Ribozyme is derived from the Hairpin Ribozyme.
Aptamers
Aptamers are RNA or DNA sequences that can selectively bind to a ligand or target. Scientists have selected a variety of aptamers for small molecules ranging from theophylline (Fig. 4A) being one of the first described aptamers
Both catalytic nucleic acids and aptamers are as for today selected via systematic evolution of ligands by exponential enhancement
It has been shown that selected aptamers can be used to make the catalytic activity dependent on the presence of a ligand.

An aptamer connected via a communication module to an catalytic sequence; The communication module translates the interaction of the ligand with the aptamer to the catalytic part of the aptazyme.

Systematic Evolution of Ligands by EXponential Enrichment (SELEX): Current method to select functional nucleic acids from a random library. Figure adapted from: Source

The Spinach Aptamer turns on fluorescence of its ligand upon bind it.
Besides the above describes functional nucleic acids SELEX revealed aptamers which not only bind to a ligand but also are able to turn on fluorescence of different non-fluorescent dyes. An interesting example for an aptamer with such a special function is the Malachite Green Aptamer.
Though we so far mainly discussed functional RNA there are also both DNA aptamers and catalytic DNA which were selected using SELEX as well. As for today the concept of functional nucleic acids in nature has only been found for RNA. Nevertheless DNA aptamers and also catalytic DNA were selected in vitro.
Catalytic DNA – DNAzymes

Secondary structure of 7-18 and 10-23 RNA-cleaving DNAzyme with substrate RNA. Cleavage site is indiacted with an arrow.
SELEX revealed a variety of DNA aptamers and catalytic DNA known as DNAzymes (or Deoxribozymes). Breaker and Joyce selected the first DNAzyme, a RNA-cleaving DNA.
To verify the prediction of the software MAWS we fused the calculated aptamers to a DNAzyme with JAWS and thus created a ligand dependent activity. For this we used the HRP-mimicking DNAzyme (Fig. 9) which was described in 1998.
Functional nucleic acids in iGEM
Looking at the history of functional RNA in iGEM naturally occurring self-splicing introns were used by Team Waterloo 2011 to rejoin a split GFP posttranscriptionally. Riboswitches the natural relatives of aptamers have been used in several projects for example Cambridge 2012, Goettingen 2013 and also Bettencourt 2015. Hammerhead ribozymes have been discovered by several teams as handy tools (Peking 2011, Fudan 2012 and MIT 2011) and DTU Denmark 2014 worked with the Spinach aptamer. Although functional RNA has been used in some iGEM projects DNA or DNAzymes have rarely been mentioned before. Our project this year aims to modularize and standardize all these and many more parts. With our new BBF RFC 110 we simplify the work with RNA and thus want to encourage future teams to also dive into the world of RNA. By the addition and modularization of many DNA parts we want to stress to possibilities DNA possesses.

HRP-mimicking DNAzyme folds into a G-quadruplex and assembles a hemin inside. IT catalyzes the reduction of H2O2 to H2O and thus lead to turn over of classical HRP substrates.
By providing the community with tools for different applications, we contribute a foundational advance in the work with functional RNA. Not only is the system highly modular and therefore predisposed for standardization and usage in Synthetic Biology, the software developed by our team enables scientists to create aptamers and aptazymes de novo. Thereby we open up the world of functional nucleic acids to the entire Synthetic Biology community. Dive deeper now!