Team:Cambridge-JIC/Stretch Goals
Screening System - A Potential use for OpenScope
The Concept
 To integrate OpenScope onto a desktop translation system (CNC), such as the Shapeoko, implementing an automated, high-throughput screening system for Enhancer Trap, Forward Mutagenesis and Reporter screens in Marchantia and potentially yeast. This involves sample scanning, detection and labelling, after macroscopic and microscopic imaging in brightfield and fluorescence.
To integrate OpenScope onto a desktop translation system (CNC), such as the Shapeoko, implementing an automated, high-throughput screening system for Enhancer Trap, Forward Mutagenesis and Reporter screens in Marchantia and potentially yeast. This involves sample scanning, detection and labelling, after macroscopic and microscopic imaging in brightfield and fluorescence.
The Approach
Using the CNC for coarse positioning (0.1mm accuracy) and OpenScope flexure-type plastic mechanisms for fine positioning (accuracy of the order of 1 μm). The Raspberry Pi camera is used for imaging. Exploit the image recognition software we have developed for labelling, and MicroMaps for image stitching to map the sample field. The original (unmodified) Raspberry Pi camera is used for macroscopic imaging, and the OpenScope optics cube (Raspberry Pi camera with inverted lens) - for microscopy.
Problems Encountered
Poor repeatability of CNC head positioning
Significant memory required for image storage
Full screening time of the order of 1 day
Theoretically, the calculations go as follows:
One image: 1024x768px, size on disk: approx. 400KB
Maximal reliable travel speed of Shapeoko head: 900cm/min
Field of view (Raspberry Pi camera with inverted lens, as in the OpenScope): approx. 50x40μm, hence area = 2000μm2=2x10-5cm2
Standard Petri dish: 90mm diameter, hence area ≈ 64cm2
Number of images required to cover whole Petri: 64cm2/2x10-5cm2≈3x106 images, i.e. 109 KB memory (approx. 1TB)
Time to scan whole area of 1 Petri: 17hrs at max speed (3x106 squares with 50μm sides); in practice constant movement with maximal speed is not possible – the head needs to start/stop/focus, so the estimated time is 24hrs or more (but this might still be decreased by a faster moving head on a more reliable CNC)
In short, after doing this math, we almost gave up on the idea. However, after some more maths, we came up with a...
Solution
Preliminary scanning can be carried out with a macroscopic (normal) camera. The image recognition software can then scan for whole colonies or Marchantia plants within the recorded images. This turns out to be much more feasible:
Camera field of view: 3.5x2.5cm, hence area ≈ 8.5cm2
Images required to cover whole Petri dish: 64cm2/8.5cm2=8 images
Accounting for overlaps required for stitching at x4 coverage (each vertex appears in 4 neighbouring images), this gives 32 images
Total memory usage: 32 x 400KB ≈ 13 MB
This approach significantly reduces the number of images and time required to cover a whole plate. When image recognition software is implemented, it is possible to label areas of interest within the whole sample field. These specific areas can then be imaged by the microscope head attachment independently.
Software Architecture

We began to develop some python libraries to automatically manage this process, with an architecture as seen in the image to the right. The software in its current state is available within the 'lib' and 'hw' directories in our Github repository, which can be found on our Downloads page (or go here to look for future revisions). It is based around a series of abstraction layers with the ultimate goal of hiding any underlying hardware and allowing easy automation of experiments. Our example use case is an automated screening system as described above, whereby a macroscopic camera images a large sample set, software is then used to identify individual samples which are independently imaged by a microscopic camera, finally these images are screened (using a range of image processing algorithms) to identify different phenotypes.
We start with the hardware itself. An xyz translation system, such as a Shapeoko or other CNC machine, is fitted with at least one 'head'. The most important head for our use is our OpenScope microscope. In our example we would also fit the Shapeoko with a macroscopic camera (or an overhead camera with a sufficiently large field of view). Further, we intend to have a marker.
The next layer up is the driver software which interacts directly with this hardware.
The Shapeoko itself is controlled by an Arduino Mega with a G-code interpreter installed which we can interface with to send complex movement commands (such as precision arcs) and to recalibrate at any time. Our driver can handle unexpected events such as cable disconnection with ease, recalibrating afterwards in case of unaccounted-for interruptions.
The OpenScope software can be directly leveraged for the camera heads.
In our calibration tests we attached a pen to the Shapeoko head. This was much simpler as we could control the z axis to apply the pen at will, without any additional Arduinos to interface with.
We also have some virtual hardware in our examples and tests to demonstrate the abilities of the software, such as a camera which can navigate a gigapixel image of the Andromeda galaxy
These drivers use a common software class to unite their differences under a common language. In this way we can abstract away from all the intricacies of each hardware head and control the hardware with ease. There are a few key classes we use here. The Head class (and Camera subclass) are used to refer to the hardware heads. The xyz stage is interacted with via the Stage class which allows for calibration, xyz movement, and head switching. Finally we unite everything under the same roof with the Workspace class. This class manages all the software commands we might send, utilising a smart queue to optimise the order in which actions are taken to reduce movement and time (and respecting the order of commands when necessary, e.g. if you’re transferring liquids with a pipette!). The workspace is the middleman through which user experiments communicate to the hardware. The last class of interest is the Canvas class. This class is responsible for collecting, and managing the stitching together of, the images that constitute an individual sample, as well as updating it periodically as necessary in case a time-lapse image is desired.
Finally comes the interesting part, your experiment! Though we don't yet have examples of this, thanks to the library abstractions provided in the lower layers, writing these experiments should be unprecedentedly easy. Python is one of the easiest languages to learn for those who are new to programming, and so should make even very complex experiments easy. See our MicroMaps page for more information on the image processing algorithms that we have been developing to characterise specimens. We have also been developing an extensive and powerful Graphical User Interface (GUI) to manage and observe your experiment, checkout our WebShell and MicroMaps interfaces under our Software section!
The automated experimental abstraction layers were developed by Will, with useful feedback and advice from the rest of the Software team.
Proof of Concept (Experiment)
Macroscopic imaging of a Petri dish containing mature Marchantia samples was carried out. Images were captured manually through the Webshell during 1-2 minutes of operation. A Raspberry Pi camera was fixed to the CNC (in our case a Shapeoko v1, 150GBP second-hand) with a 3D-printed static mount. The motorised z-axis driven by the python program allowed for focusing of the sample. Seven images were successfully stitched, in a remarkable stitching time of 1.5sec. It is notable that the stitching algorithm copes well even with frames which are rotated at a small angle relative to each other.
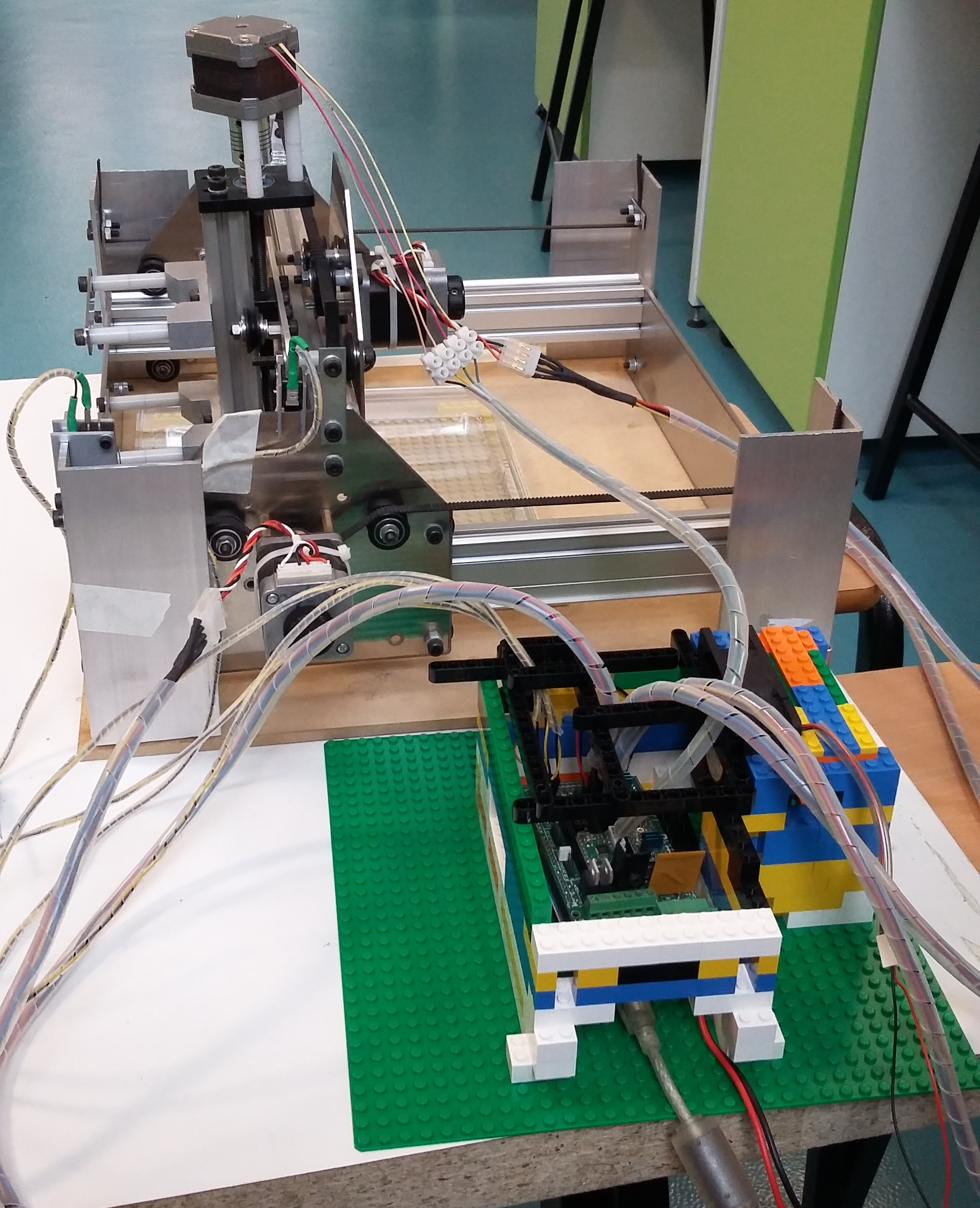
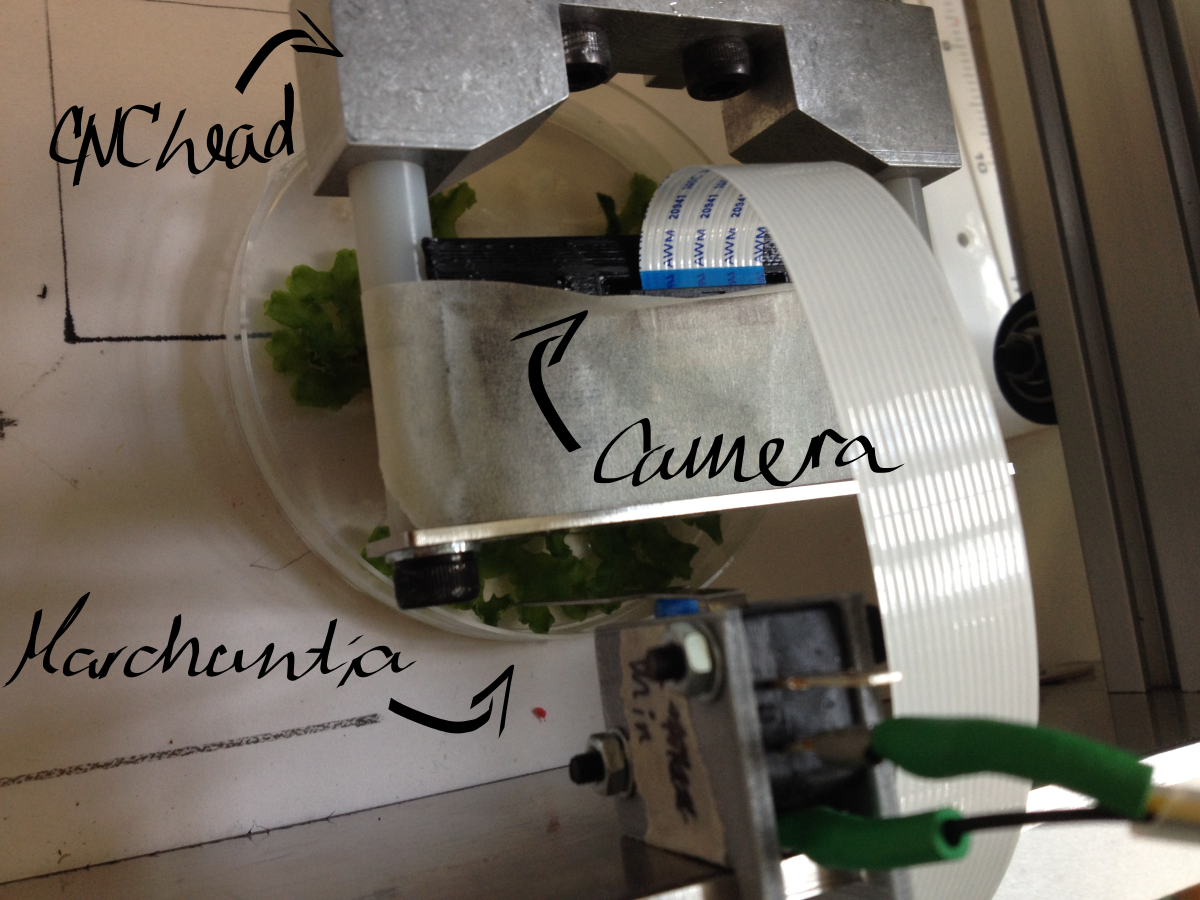

Future opportunities
Incorporate of OpenScope onto the CNC head.
Develop colony screening for yeast/bacterial colonies on Petri dishes. Criteria such as distinct colour or fluorescence can be incorporated into the image recognition software. However, the fluorescence imaging achieved on OpenScope is not currently reliable enough to perform fluorescence screening.
Implement focus-stacking software for 3D samples (like Marchantia). This facilitates the development of programs which track the growth of samples.
ABOUT US
We are a team of Cambridge undergraduates, competing in the Hardware track in iGEM 2015.
read moreLOCATION
Department of Plant Sciences,
University of Cambridge
Downing Street
CB2 3EA
CONTACT US
Email: igemcambridge2015@gmail.com
Tel: +447721944314