Difference between revisions of "Team:Cambridge-JIC/Stretch Goals"
Simonhkswan (Talk | contribs) |
Simonhkswan (Talk | contribs) |
||
| Line 7: | Line 7: | ||
<br> | <br> | ||
<h3>The Concept</h3> | <h3>The Concept</h3> | ||
| − | <p><img src="https://static.igem.org/mediawiki/2015/e/ea/CamJIC-CNC_system_b%26w.png" style="width:300px; float:right; margin-left: 10px">To integrate OpenScope onto a desktop translation system (CNC), such as the <a href="http://www.shapeoko.com/" class="blue">Shapeoko</a>, implementing an automated, high-throughput screening system for Enhancer Trap, Forward Mutagenesis and Reporter screens in <i>Marchantia</i> and potentially yeast. This | + | <p><img src="https://static.igem.org/mediawiki/2015/e/ea/CamJIC-CNC_system_b%26w.png" style="width:300px; float:right; margin-left: 10px">To integrate OpenScope onto a desktop translation system (CNC), such as the <a href="http://www.shapeoko.com/" class="blue">Shapeoko</a>, implementing an automated, high-throughput screening system for Enhancer Trap, Forward Mutagenesis and Reporter screens in <i>Marchantia</i> and potentially yeast. This involves sample scanning, detection and labelling, after macroscopic and microscopic imaging in brightfield and fluorescence.</p> |
<h3>The Approach</h3> | <h3>The Approach</h3> | ||
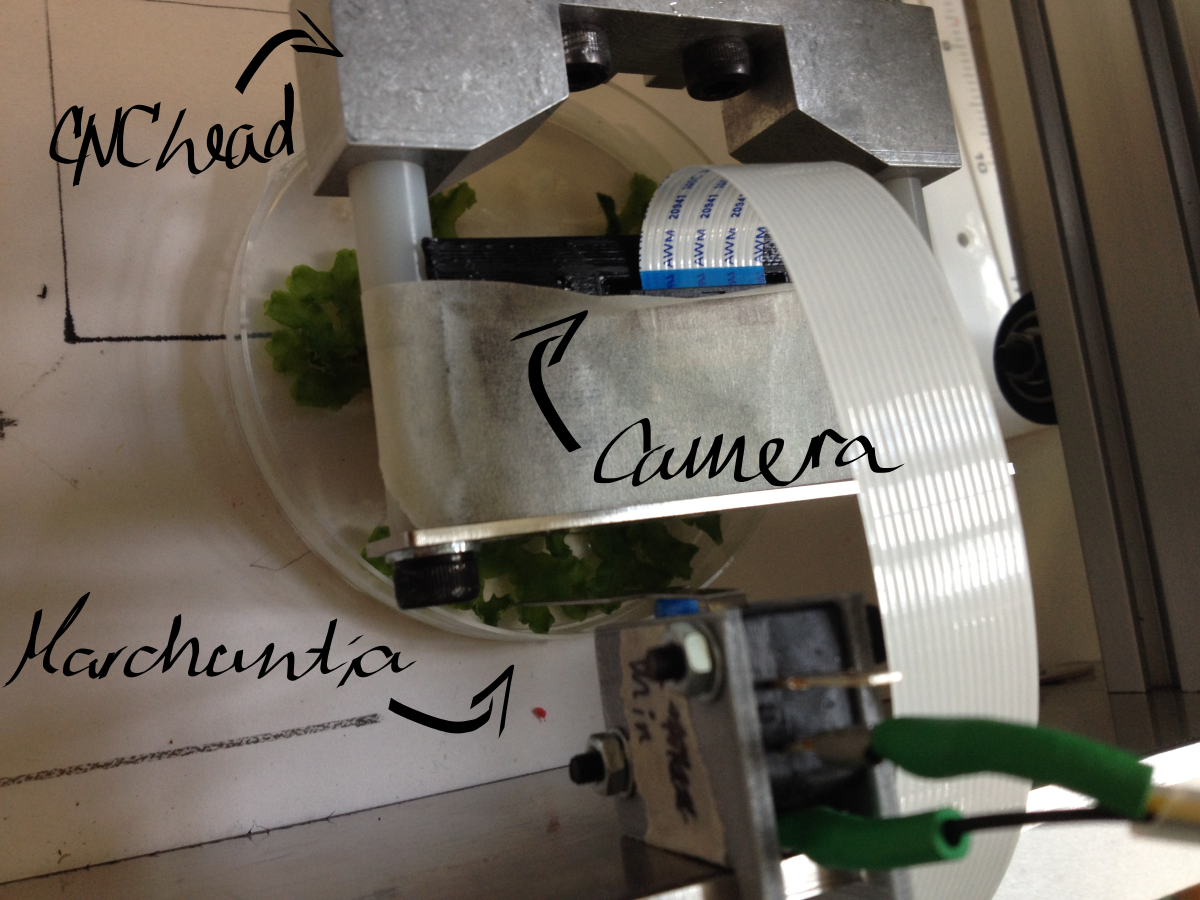
| − | <p>Using the CNC for coarse positioning (0.1mm accuracy) and OpenScope flexure-type plastic mechanisms for fine positioning (accuracy of the order of 1 μm). The Raspberry Pi camera | + | <p>Using the CNC for coarse positioning (0.1mm accuracy) and OpenScope flexure-type plastic mechanisms for fine positioning (accuracy of the order of 1 μm). The Raspberry Pi camera is used for imaging. Exploit the image recognition software we have developed for labelling, and <a href="//2015.igem.org/Team:Cambridge-JIC/MicroMaps" class="blue">MicroMaps</a> for image stitching to map the sample field. The original (unmodified) Raspberry Pi camera is used for macroscopic imaging, and the OpenScope optics cube (Raspberry Pi camera with inverted lens) - for microscopy.</p> |
<h3>Software Development</h3> | <h3>Software Development</h3> | ||
| − | <p>A Python driver script was written to control the Shapeoko. The code allows for | + | <p>A Python driver script was written to control the Shapeoko. The code allows for a variety of actions: homing, positioning, collision feedback, speed control, path following, etc. The <a href="//2015.igem.org/Team:Cambridge-JIC/Webshell" class="blue">Webshell</a> interface with OpenScope can be used for imaging and <a href="//2015.igem.org/Team:Cambridge-JIC/MicroMaps" class="blue">MicroMaps</a> for processing.</p> |
<h3>Problems Encountered</h3> | <h3>Problems Encountered</h3> | ||
<ul> | <ul> | ||
| Line 23: | Line 23: | ||
<li><p>Maximal reliable travel speed of Shapeoko head: 900cm/min</p></li> | <li><p>Maximal reliable travel speed of Shapeoko head: 900cm/min</p></li> | ||
<li><p>Field of view (Raspberry Pi camera with inverted lens, as in the OpenScope): approx. 50x40μm, hence area = 2000μm<sup>2</sup>=2x10<sup>-5</sup>cm<sup>2</sup></p></li> | <li><p>Field of view (Raspberry Pi camera with inverted lens, as in the OpenScope): approx. 50x40μm, hence area = 2000μm<sup>2</sup>=2x10<sup>-5</sup>cm<sup>2</sup></p></li> | ||
| − | <li><p>Standard Petri dish: 90mm diameter | + | <li><p>Standard Petri dish: 90mm diameter, hence area ≈ 64cm<sup>2</sup><p></li> |
<li><p>Number of images required to cover whole Petri: 64cm<sup>2</sup>/2x10<sup>-5</sup>cm<sup>2</sup>≈3x10<sup>6</sup> images, i.e. 10<sup>9</sup> KB memory (approx. 1TB)</p></li> | <li><p>Number of images required to cover whole Petri: 64cm<sup>2</sup>/2x10<sup>-5</sup>cm<sup>2</sup>≈3x10<sup>6</sup> images, i.e. 10<sup>9</sup> KB memory (approx. 1TB)</p></li> | ||
<li><p>Time to scan whole area of 1 Petri: 17hrs at max speed (3x10<sup>6</sup> squares with 50μm sides); in practice constant movement with maximal speed is not possible – the head needs to start/stop/focus, so the estimated time is 24hrs or more (but this might still be decreased by a faster moving head on a more reliable CNC)</p></li> | <li><p>Time to scan whole area of 1 Petri: 17hrs at max speed (3x10<sup>6</sup> squares with 50μm sides); in practice constant movement with maximal speed is not possible – the head needs to start/stop/focus, so the estimated time is 24hrs or more (but this might still be decreased by a faster moving head on a more reliable CNC)</p></li> | ||
| Line 29: | Line 29: | ||
<p><i>In short, after doing this math, we almost gave up on the idea. However, after some more maths, we came up with a...</i></p> | <p><i>In short, after doing this math, we almost gave up on the idea. However, after some more maths, we came up with a...</i></p> | ||
<h3>Solution</h3> | <h3>Solution</h3> | ||
| − | <p>Preliminary scanning can be carried out with a macroscopic (normal) camera. The image recognition software | + | <p>Preliminary scanning can be carried out with a macroscopic (normal) camera. The image recognition software can then scan for whole colonies or <i>Marchantia</i> plants within the recorded images. This turns out to be much more feasible:</p> |
<ul> | <ul> | ||
<li><p>Camera field of view: 3.5x2.5cm, hence area ≈ 8.5cm<sup>2</sup></p></li> | <li><p>Camera field of view: 3.5x2.5cm, hence area ≈ 8.5cm<sup>2</sup></p></li> | ||
| Line 38: | Line 38: | ||
<p>This approach significantly reduces the number of images and time required to cover a whole plate. When image recognition software is implemented, it is possible to label areas of interest within the whole sample field. These specific areas can then be imaged by the microscope head attachment independently.</p> | <p>This approach significantly reduces the number of images and time required to cover a whole plate. When image recognition software is implemented, it is possible to label areas of interest within the whole sample field. These specific areas can then be imaged by the microscope head attachment independently.</p> | ||
<h3>Proof of Concept (Experiment)</h3> | <h3>Proof of Concept (Experiment)</h3> | ||
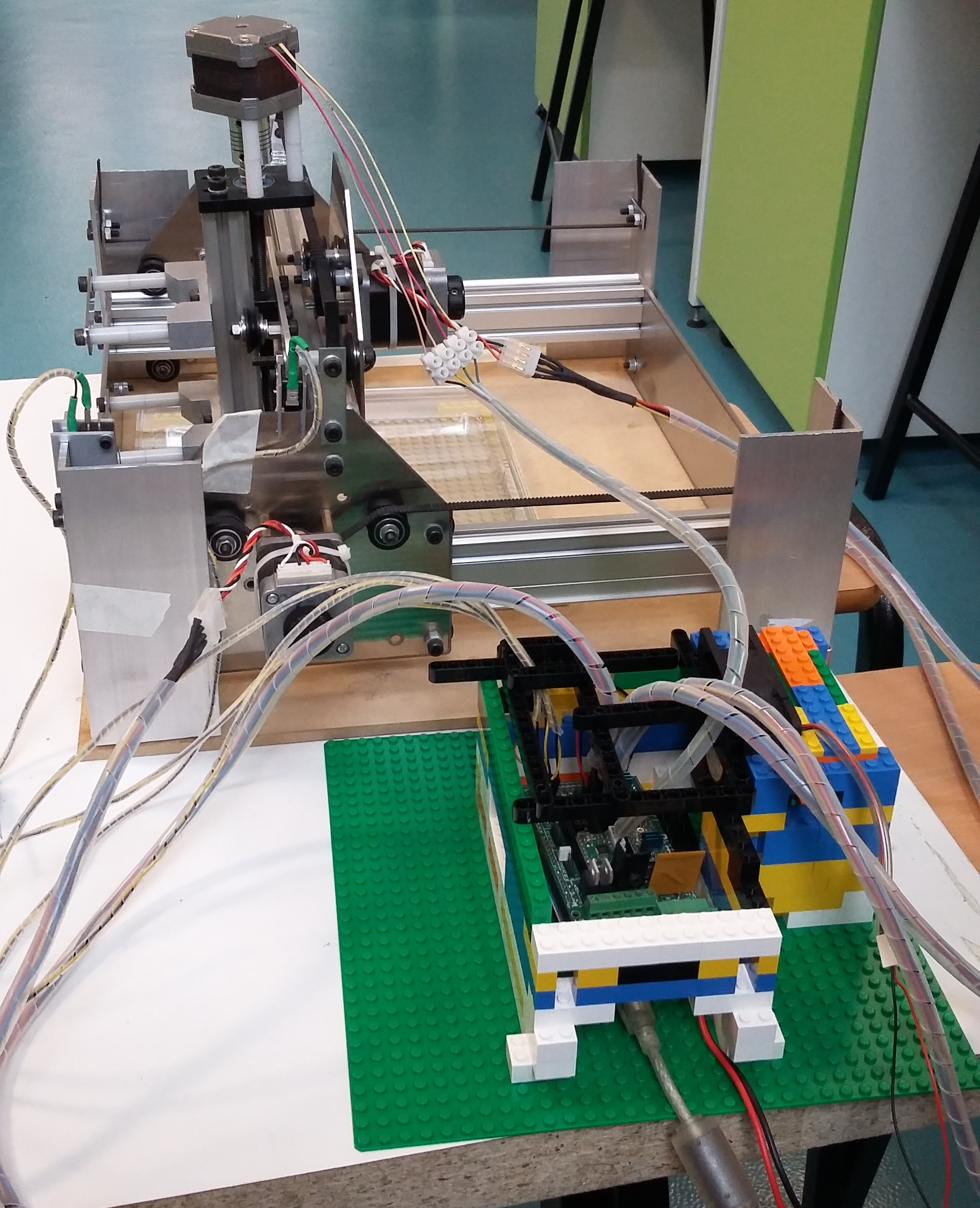
| − | <p>Macroscopic imaging of a Petri dish containing mature <i>Marchantia</i> samples was carried out. Images were captured manually through the Webshell during 1-2 minutes of operation. A Raspberry Pi camera was fixed to the CNC (in our case a Shapeoko v1, 150GBP second-hand) with a 3D-printed static mount. The motorised z-axis driven by the python program allowed for focusing of the sample. Seven images were successfully stitched, in a remarkable stitching time of 1.5sec. It is notable that the stitching algorithm copes well | + | <p>Macroscopic imaging of a Petri dish containing mature <i>Marchantia</i> samples was carried out. Images were captured manually through the Webshell during 1-2 minutes of operation. A Raspberry Pi camera was fixed to the CNC (in our case a Shapeoko v1, 150GBP second-hand) with a 3D-printed static mount. The motorised z-axis driven by the python program allowed for focusing of the sample. Seven images were successfully stitched, in a remarkable stitching time of 1.5sec. It is notable that the stitching algorithm copes well even with frames which are rotated at a small angle relative to each other. |
<center><img src="//2015.igem.org/wiki/images/9/92/CamJIC-StretchGoals-Shapeoko.png" style="height:270px;margin:10px"><img src="//2015.igem.org/wiki/images/8/8b/CamJIC-StretchGoals-Experiment.png" style="height:270px;margin:10px"><img src="//2015.igem.org/wiki/images/f/f7/CamJIC-StretchGoals-MarchantiaStitch.png" style="height:270px;margin:10px"></center></p> | <center><img src="//2015.igem.org/wiki/images/9/92/CamJIC-StretchGoals-Shapeoko.png" style="height:270px;margin:10px"><img src="//2015.igem.org/wiki/images/8/8b/CamJIC-StretchGoals-Experiment.png" style="height:270px;margin:10px"><img src="//2015.igem.org/wiki/images/f/f7/CamJIC-StretchGoals-MarchantiaStitch.png" style="height:270px;margin:10px"></center></p> | ||
<p><center><i>Desktop translation system (CNC) Shapeoko v.1, the setup used for imaging and resultant stitched image</i></center></p> | <p><center><i>Desktop translation system (CNC) Shapeoko v.1, the setup used for imaging and resultant stitched image</i></center></p> | ||
<h3>Future opportunities</h3> | <h3>Future opportunities</h3> | ||
| − | <p> | + | <p>Incorporate of OpenScope onto the CNC head.</p> |
<p>Develop colony screening for yeast/bacterial colonies on Petri dishes. Criteria such as distinct colour or fluorescence can be incorporated into the image recognition software. However, the fluorescence imaging achieved on OpenScope is not currently reliable enough to perform fluorescence screening.</p> | <p>Develop colony screening for yeast/bacterial colonies on Petri dishes. Criteria such as distinct colour or fluorescence can be incorporated into the image recognition software. However, the fluorescence imaging achieved on OpenScope is not currently reliable enough to perform fluorescence screening.</p> | ||
| − | <p>Implement focus-stacking software for 3D samples (like <i>Marchantia</i>). This | + | <p>Implement focus-stacking software for 3D samples (like <i>Marchantia</i>). This facilitates the development of programs which track the growth of samples.</p> |
</div> | </div> | ||
</div> | </div> | ||
Revision as of 18:57, 15 September 2015
 To integrate OpenScope onto a desktop translation system (CNC), such as the
To integrate OpenScope onto a desktop translation system (CNC), such as the