Team:UCSF/Results

HOVER TOENLARGE
EXPERIMENTS/RESULTS
PROJECT ACHIEVEMENTS
- We built a communication circuit in yeast that exhibits stable divergent populations after communication even though their responses to an initial stimulus are similar.
- We characterized several communication parameters to further optimize this output.
- We submitted and characterized BioBrick parts of almost all components of our circuit.
EXPERIMENTAL DESIGN
Our goal for our circuit is to elicit a divergent community response from two genetically similar cells by varying different communication parameters. To measure distinct community responses in our circuit, we will look at INDIVIDUAL (GFP) response versus COMMUNITY (RFP) response.


We quantify our readouts by inducing our genetic circuit with varying concentrations of doxycycline, our stimulus, and measuring the fluorescent readouts with flow cytometry at 0, 1.5, 3, and 5 hour timepoints.

BASIC CIRCUIT TESTING
To test the functionality of our basic circuit, we characterized and validated the fluorescent readouts for individual and community response to ensure they would provide accurate and robust measurements. To do so, we ran three different experiments:
- Characterization of our Community Response Readout
- Validation of LexA Transcription Factor Feedback Sensitivity
- Stimulus Induction of Basic Circuit with Altered Communication Parameters
1. Characterization of Community Response Readout:
In our circuit design, our community response is read out by RFP fused with a LexA transcription factor chimera. This readout is regulated by an alpha factor responsive promoter (αFRP), either pFig2c or pAga1. It is vital that these αFRPs have a high dynamic range to distinguish between different alpha factor concentrations and sense a difference between cell populations (i.e. activated vs. unactivated).

Our team characterized pFig2c (BBa_K1829002) and pAga1 (BBa_K1829005) with an alpha factor dose response to measure sensitivity to the community signal. Our testing supports that pAga1 is a better αFRP to use to measure community response in our circuit due to greater dynamic range and a higher expression peak.

2. Validation of LexA Transcription Factor Feedback Sensitivity:
Our project this year is aimed toward changing communication parameters to elicit divergent behavior. We’ve engineered these communication parameters into our yeast under the regulation of a LexA transcription factor inducible promoter, LexAOps. This transcription factor is a synthetic fusion of the DNA-binding domain of the repressor LexA with transcription activation domains from VP64 (BBa_J176013). We control production of the LexA-TF with an alpha factor inducible promoter, and when present it binds to the LexA operator sequence to induce transcription of downstream genes. Thus, when our cells sense alpha factor, they will amplify the signal to produce some sort of positive feedback, such as increased secretion or increased reception. (See Circuit design for more detail). Because these feedback loops are essential to our desired phenotype, we constructed a strain to measure the sensitivity of our feedback loops to alpha factor concentrations.

We induced our yeast strain with varying concentrations of purified alpha factor and recorded the cells' level of florescence after two hours. Our experiments verified that our feedback loops were sensitive to alpha factor.

3. Stimulus Induction of Basic Circuit with Altered Communication Parameters:
Now that we have validation that our basic circuit is functional and that the genetic components fulfill their intended purposes, we are ready to begin testing our strains for divergent community phenotypes. We tested our “basic” circuit, containing only the reporters and communication signal in comparison to a Positive Feedback circuit, which produces both mating factor alpha and the receptor Ste2 upon sensing of alpha factor. To test these circuits, we induce our genetic circuit with our stimulus, doxycycline, and measure individual and community readouts with flow cytometry.

Our experiments show genetically identical cells with high individual responses participating in bimodal activation when utilizing a positive feedback loop.We have achieved our goal of creating a population of genetically identical cells that diverge into phenotypically distinct cells.

Although we achieved our goal of creating divergent populations from genetically identical cells, we ran into a major problem in our circuit: leaky expression. Because we are working with Bar1 knockout yeast strains, our circuit is hypersensitive to alpha factor signal. So the slightest reception of signal will trigger positive feedback and propagate activation. We see leaky expression in our circuit that activates our circuit at 0 hours, most likely in the pTET system (which drives production of GFP and alpha factor signal). We can also see high basal levels of expression in GFP plots as well.

CREATING AN OPTIMAL CIRCUIT
In order to improve the functionality of our circuit, we attempted to control the amount of alpha factor being sensed and secreted through two means:
1. Signal Degradation:
We believe that implementing Bar1, a protease that degrades alpha factor, will work in reducing leakiness. We have constructed motifs that express Bar1 under the pTEF1 constitutive promoter and its four mutants characterized by UCSF iGEM 2014.

Thus, we have an array of signal degradation levels which we can implement into our circuit. Previous work has shown Bar1 as an integral mechanism of yeast to desensitize them to alpha factor and sharpen concentration gradients while mating [3]. We hypothesize that Bar1 will create a threshold for activation by degrading background alpha factor secreted into the media. Therefore, only cells with strong enough positive feedback will be able to sense alpha factor and activate a community response. To test this, we measured the expression of an alpha factor responsive promoter under conditions without Bar1 and with Bar1. This experiment verified Bar1’s ability to desensitize yeast to alpha factor and polarize the gaps between “ON” and “OFF.”

However, when testing this motif in our circuit, we discovered that Bar1 was too strong at all levels (low to high) for all yeast cells in the population to overcome. This meant that our lowest expressing Bar1 construct was still too effective at degrading alpha factor to allow any cell in the population to activate. For a more feasible activation threshold, we would need to regulate Bar1 with a weaker expressing promoter.

2. Spatial Clustering:
We believe that spatial retention of cells will stabilize the divergence we see in our population. We have constructed motifs that express a fusion yeast surface display protein, Mgfp5.
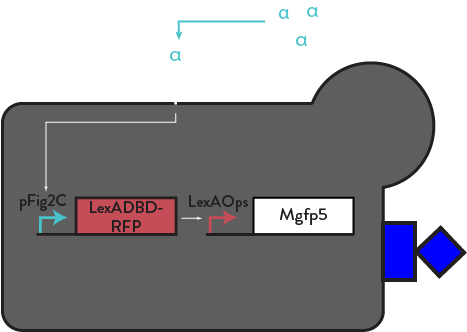
This protein is endogenously found in mussel feet to secrete a bioadhesive that allows them to stick to rocks. We have engineered this protein to be displayed on the cell surface of yeast, and only expressed by activated cells, through a similar positive feedback motif as Ste2 and mFα. Activated cells will cluster and form a smaller, exclusive community and increase their own activation while not engaging with unactivated cells (who remain silent). Similar models are utilized by T-Cells through lymph nodes [9], and V. fischeri through symbiotic organs in squids [12] in order to communicate with their community more efficiently. Once in a cluster, cells will amplify a local concentration of alpha factor and participate in asocial, exclusive behavior [3]. This motif should lend to a more stable, replicable, and efficient method of eliciting bimodal activation. We have constructed and implemented this clustering motif (LexAOps-Mgfp5) into our basic circuit with positive feedback. We have done some preliminary qualitiative testing, but hope to proceed with quantitative testing of cluster size with software such as CellProfiler or ImageJ in the near future. With visual analysis using light microscopy, we observed large clusters after induction with alpha factor.

FUTURE DIRECTION

Although we have achieved our goal of eliciting bimodal activation in our yeast cell populations, we hope to optimize this system to create a more stable and robust divergent response. The obvious next step is to work to tune the Bar1 level to both reduce background/set a threshold and allow for activation. We characterized the activity of the endogenous promoter for Bar1 (pBar1, BBa_K1829001), and found that it exhibits little to no activation when induced with alpha factor. Based on the overwhelming activity of Bar1 in our circuit, we suspect that pBar1 has low activity in order to prevent production of too much Bar1. Perhaps with less Bar1 we can create more robust divergent populations in our community. Alternatively, increasing the strength of positive feedback -- either by spatially promoting it through clustering or altering the components of the feedback portion to express at higher levels -- could help overcome Bar1 degradation to induce activation of the circuit. In addition to divergent behaviors, we still hope to model more complex community phenotypes. Other communication parameters we could look at could be the temporal dynamics of activation and paracrine vs. autocrine signaling. Temporal dynamics are crucial to look at to understand the efficiency and timescale of activation in cellular populations. For example, time is an important aspect in T cell proliferation as extended activation can result in autoimmune disorders. So populations must somehow be able to tune temporal scales and scale back activation after the response is no longer necessary. As a necessary step in looking in dynamics, we may want to alter the system to optimize our reporters. Because RFP is a stable protein, expression of RFP will degrade slowly and stay in the system, offering remnant readouts from previous time points. By creating a less stable community response readout, we can eliminate this artifact in our data. This time-dependent community response will then provide new insights into how activation is started, maintained, and degraded over time. Ultimately, we hope that by building a model system for studying how cell-to-cell communication can help process responses of a community of cells to a stimulus, we might learn more about how to perturb or harness natural systems to produce new synthetic biology systems, tools, or therapeutics.
