Difference between revisions of "Team:HKUST-Rice/Application"
Margaret.lie (Talk | contribs) |
Margaret.lie (Talk | contribs) |
||
| Line 71: | Line 71: | ||
We quickly realized that in order to deploy our NPK sensors across multiple strains of soil bacteria, we would need to build the genetic circuitry on broad-host range plasmids. Before putting our sensors into soil bacteria, we decided to show proof-of-concept by using a conjugation plasmid with an RP4 transfer system, green fluorescent protein (GFP), and a chloramphenicol selectable marker. The GFP serves as a visual output. </p> | We quickly realized that in order to deploy our NPK sensors across multiple strains of soil bacteria, we would need to build the genetic circuitry on broad-host range plasmids. Before putting our sensors into soil bacteria, we decided to show proof-of-concept by using a conjugation plasmid with an RP4 transfer system, green fluorescent protein (GFP), and a chloramphenicol selectable marker. The GFP serves as a visual output. </p> | ||
<p> | <p> | ||
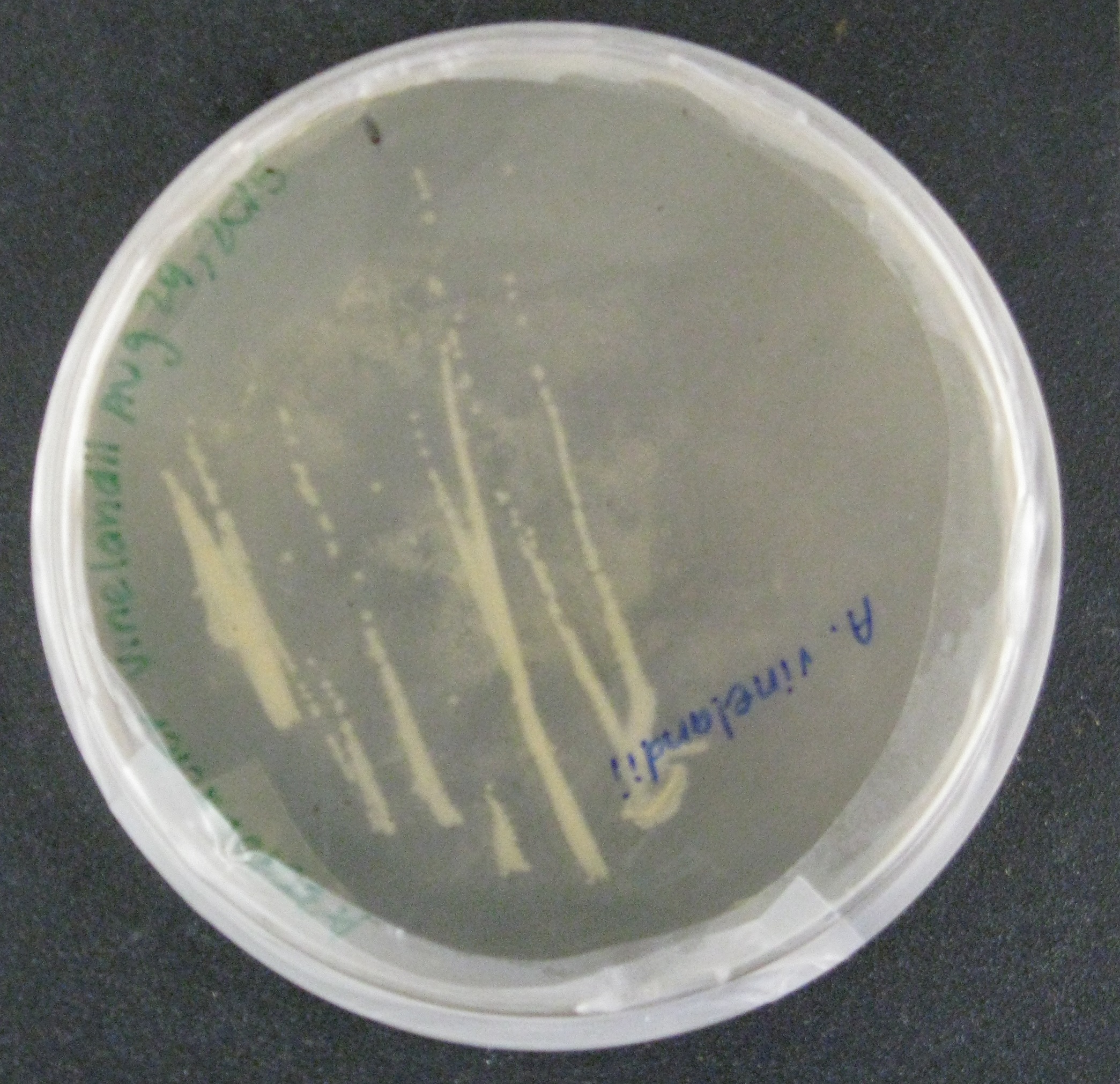
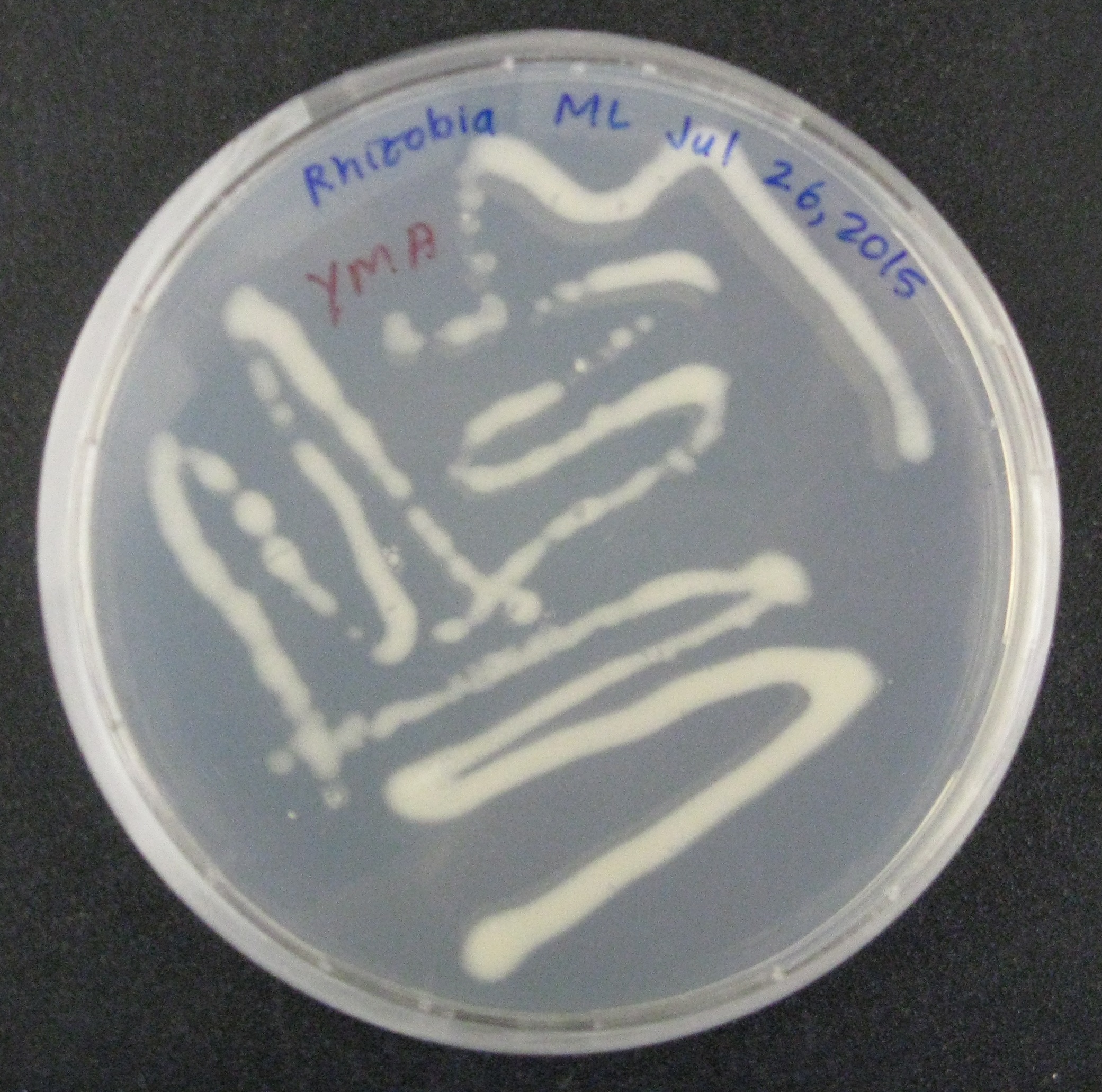
| − | We then began doing proof-of-concept experiments to transform our broad-host range plasmid containing GFP into two strains of gram-negative soil bacteria: <i>Rhizobia leguminosarum</i>, a nitrogen-fixing microbe, was conjugated to <i>E. coli</i> S17, which along with the conjugation plasmid, provided all the genes necessary to transfer the GFP containing plasmid into the soil bacteria. Additionally, <i>Azotobacter vinelandii</i>, another nitrogen-fixing bacteria, was transformed by electroporation using the conjugation plasmid. We were successful in getting the plasmid into both species. | + | We then began doing proof-of-concept experiments to transform our broad-host range plasmid containing GFP into two strains of gram-negative soil bacteria: <i>Rhizobia leguminosarum</i>, a nitrogen-fixing microbe, was conjugated to <i>E. coli</i> S17, which along with the conjugation plasmid, provided all the genes necessary to transfer the GFP containing plasmid into the soil bacteria. Additionally, <i>Azotobacter vinelandii</i>, another nitrogen-fixing bacteria, was transformed by electroporation using the conjugation plasmid. We were successful in getting the plasmid into both species. </p> |
<img src="https://static.igem.org/mediawiki/2015/5/5f/Table_Rice.png"> | <img src="https://static.igem.org/mediawiki/2015/5/5f/Table_Rice.png"> | ||
Revision as of 02:44, 18 September 2015
Application
After designing NPK biosensors and talking with farmers about their needs, we brainstormed potential applications for our system of genetic circuits.
At its inception, the goal of this project was to design a method of determining macronutrient concentrations that was cheap, simple, and easy to use for farmers around the world. However, when we did research on what products were currently available to sense soil macronutrient concentrations, we concluded that many satisfactory, low-cost chemical assays have already been developed. For example, there is a Ferry-Morse Soil Test Kit available for $17 at The Home Depot and universities like Texas A&M offer lab soil tests for $10 per sample.
We decided to explore where the current NPK tests could be improved, so we listed out a few of the most essential criteria for design and deployment of a sensor. A good sensor for this application is accurate, easy to use, low cost, gives a quick readout of the results, and is composed of relatively stable components. Examining the NPK assays currently available, we found that while most provide cheap and rapid measurement of NPK levels, they do so at the cost of accuracy. We designed the biosensors to be useful registry parts suitable for many future applications and did not specifically design them to improve accuracy. However, we would still like to test if the biological, rather than purely chemical, aspect of our engineered biosensors enables them to get a more accurate reading of what nutrients are readily available from the perspective of a growing plant.
Considering the accessibility of chemical NPK assays and public opinion toward biosensors, we have begun developing two separate applications of the NPK sensors: a paper-based lysate and a controlled biofertilizer system.
Paper-based Lysate
We want to deploy our sensors in a way that would alleviate biosafety concerns and continue to be quick and low-cost so that it continues to be competitive with current products. With these criteria in mind, we decided to make a paper-based testing assay. We aim to create a freeze-dried cell-free system on paper, based on the research of Pardee, et. al, that will be easy to use (Pardee, et. al, 2014). Using a liquid TX-TL kit ordered from promega and the associated protocol, we lysed E. coli expressing mCherry (Protocol). We measured a negative control, positive control, and our sample on a fluorescence plate reader and saw that The mCherry strain had higher fluorescence than the negative control or a sample expressing luciferase. Similarly, a luminescence assay revealed significant levels for luciferase, and very low levels for the mCherry sample and the negative control. This data was collected on a tecan plate reader.


The next step is to move our NPK biosensors into this cell-free system, and then move from liquid cell lysate to a paper-based system. That way, our sensors can be easily used by farmers without biosafety concerns. The idea is that the paper-based system can be rehydrated water that has been used to dilute a soil sample. Then, different colorimetric outputs will be produced by the system depending on the different NPK levels detected in the soil sample.
Controlled Biofertilizer System
In the future, we envision an application that would completely remove the need for farmers to constantly test and fertilize their soil. Our second application is a system in which a modified soil-bacterium can sense the concentration of a macronutrient in its environment, and then make a decision to add or remove that nutrient from the soil as needed.
Biofertilizer has been defined as the use of microorganisms to increase the availability and uptake of mineral nutrients for plants (Vessey, 2003). Depending on the crop, native soil species of nitrogen-fixing, denitrifying, phosphate-solubilizing, and potassium-solubilizing bacteria have been identified (Vessey, 2003, Ngoc Diep & Ngoc Hieu, 2014). By attaching our individual nutrient-sensors to the solubilizing- or consumption-machinery of each of these bacterial species, you could theoretically control the increase or decrease that nutrient’s concentration and availability in the soil, depending on the output of the sensor.
We quickly realized that in order to deploy our NPK sensors across multiple strains of soil bacteria, we would need to build the genetic circuitry on broad-host range plasmids. Before putting our sensors into soil bacteria, we decided to show proof-of-concept by using a conjugation plasmid with an RP4 transfer system, green fluorescent protein (GFP), and a chloramphenicol selectable marker. The GFP serves as a visual output.
We then began doing proof-of-concept experiments to transform our broad-host range plasmid containing GFP into two strains of gram-negative soil bacteria: Rhizobia leguminosarum, a nitrogen-fixing microbe, was conjugated to E. coli S17, which along with the conjugation plasmid, provided all the genes necessary to transfer the GFP containing plasmid into the soil bacteria. Additionally, Azotobacter vinelandii, another nitrogen-fixing bacteria, was transformed by electroporation using the conjugation plasmid. We were successful in getting the plasmid into both species.

We hope to confirm our findings further by amplifying the nitrogenase gene, only present in R. leguminosarum and A. vinelandii. Alternative studies utilizing the plate reader will analyze the fluorescence of the GFP.


While this application of our NPK sensors is still very much in the initial stages of development, we are very excited by the idea of nutrient-controlled biofertilizer bacteria. This project fully utilizes the potential biocomputing power that can be introduced into bacteria and we hope to continue developing it future years. We envision a day when we resolve biosafety issues and farmers will no longer have to test their soil, determine nutrient deficiencies, and correct them with fertilizers. Instead, our genetically modified soil-bacteria living with the crops will continuously test and replenish or destroy nitrogen, phosphate, and potassium, depending on the specific requirements of the crop. This system would not only make farming easier, by eliminating the need to add fertilizer, it would prevent runoff and have a major positive environmental impact.
References
Ferry Morse 40 Test Soil Test Kit. (n.d.). Retrieved from http://www.homedepot.com/p/Ferry-Morse-40-Test-Soil-Test-Kit-920/202819850
Soil Sample Information Form. (n.d.). Retrieved from http://soiltesting.tamu.edu/files/soilwebform.pdf
Pardee, K., Green, A., Ferrante, T., Cameron, D., Daley-Keyser, A., Yin P., Collins, J. (2014). Paper-Based Synthetic Gene Networks. Cell, 159(4), 940-954.
Vessey, K.J. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant and Soil, 255(2), 571-586.
Ngoc Diep, C., & Ngoc Hieu, T. (2013). Phosphate and potassium solubilizing bacteria from weathered materials of denatured rock mountain, Ha Tien, Kiên Giang province, Vietnam. American Journal of Life Sciences, 3(1), 88-92

