Difference between revisions of "Team:UCLA/Notebook/Honeybee Silk"
(→Cloning) |
(→Cloning) |
||
| (36 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
{{:Team:UCLA/CSS}} | {{:Team:UCLA/CSS}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | =HONEYBEE SILK PROJECT= | ||
| Line 368: | Line 11: | ||
==<u>Achievements</u>== | ==<u>Achievements</u>== | ||
| − | As of | + | As of 9/18 we are well on our way to establishing a strong foundation for this project. We have cloned five honey bee sequences into biobrick approved format, have expressed and purified honey bee silk protein. (For detailed achievements see below) |
===Cloning=== | ===Cloning=== | ||
| − | We have successfully cloned and sequence verified ''' | + | We have successfully cloned and sequence verified '''five''' honeybee silk constructs into the igem psb1c3 plasmid backbone, in the correct biobrick format. A comprehensive list of our biobrick parts can be found [https://2015.igem.org/Team:UCLA/Parts here] |
| − | *The first sequence is just the honeybee silk protein #3 in the psb1c3 backbone. The annotated sequence, along with further details on how we designed and cloned this biobrick can be found here. [http://parts.igem.org/Part:BBa_K1763000 BBa_K1763000] | + | *The first sequence is just the honeybee silk protein #3 in the psb1c3 backbone. The annotated sequence, along with further details on how we designed and cloned this biobrick can be found here. [http://parts.igem.org/Part:BBa_K1763000 BBa_K1763000] |
| − | *The second sequence is the same silk coding sequence, but with regulatory elements upstream for protein expression, including [http://parts.igem.org/Part:BBa_R0010 BBa_R0010 Promoter] and [http://parts.igem.org/Part:BBa_B0034 RBS BBa_B0034 ]. The sequence can be found here. [ | + | *The second sequence is the same silk coding sequence, but with regulatory elements upstream for protein expression, including [http://parts.igem.org/Part:BBa_R0010 BBa_R0010 Promoter] and [http://parts.igem.org/Part:BBa_B0034 RBS BBa_B0034 ]. The sequence can be found here. [http://parts.igem.org/Part:BBa_K1763001 BBa_K1763001 ] along with further details on the design and cloning of this biobrick. |
| + | *The third sequence is a fusion protein between our honeybee silk and the Spycatcher, protein which has specific affinity for SpyCatcher. It also contains regulatory elements like a promoter and rbs for expression. You can view the sequence [http://parts.igem.org/Part:BBa_K1763008 BBa_K1763008 here .] | ||
| + | *The fourth sequence [http://parts.igem.org/Part:BBa_K1763007 BBa_K1763007 ] is our honeybee silk sequence with a T7 promoter upstream for effective protein expression in bacterial strains containing t7 polymerase. | ||
| + | *The fifth sequence, [http://parts.igem.org/Part:BBa_K1763015 BBa_K1763015] is our honeybee silk sequence+sfGFP with a T7 promoter and RBS upstream that can be used in bacterial strains that contain T7 polymerase. | ||
===Protein Expression=== | ===Protein Expression=== | ||
| − | + | *We have just finished our first round of honey bee silk expression from [http://parts.igem.org/Part:BBa_K1763001 BBa_K1763001] | |
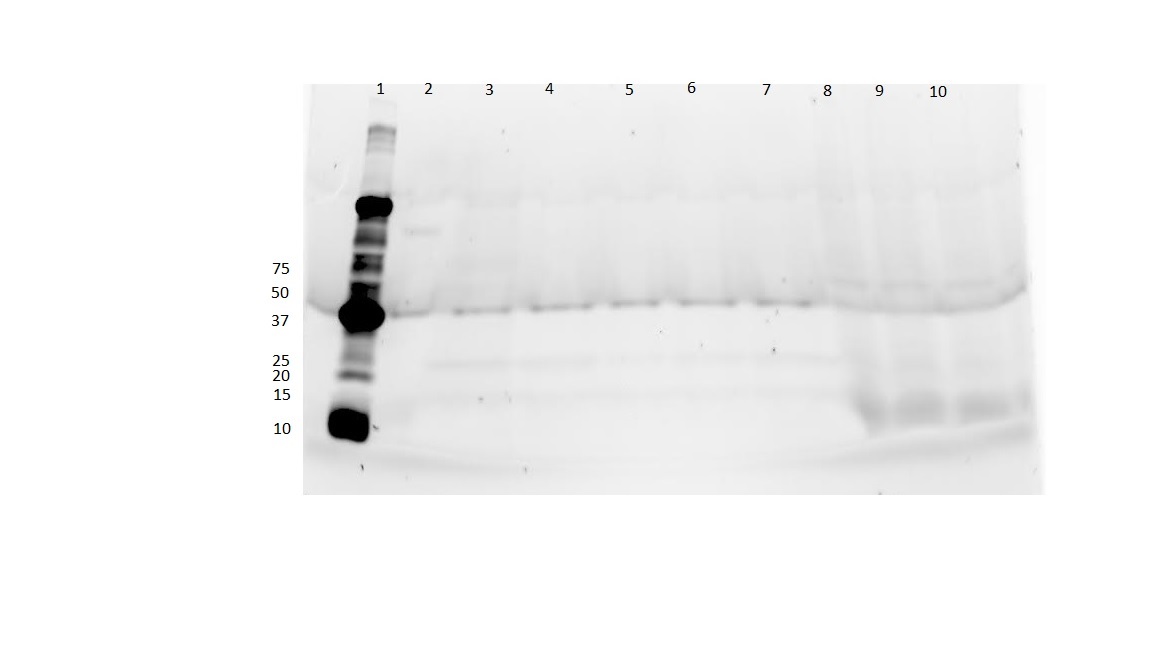
| + | *[https://2015.igem.org/Team:UCLA/Notebook/Honeybee_Silk/26_May_2015 Here] is our first attempt at protein expression along with the gel. | ||
| + | [[File:UCLAHoneybee.jpg|none|thumb|300px|'''Fig. 1''' Expected size of product is 40.0 kDA]] | ||
| + | |||
| + | *Unfortunately the purified protein that we see on the gel (should be in next to last lane on the right) doesn't seem to be the right size. | ||
| + | **We will try again using a T7 promoter and a slightly modified protein expression and purification protocol. | ||
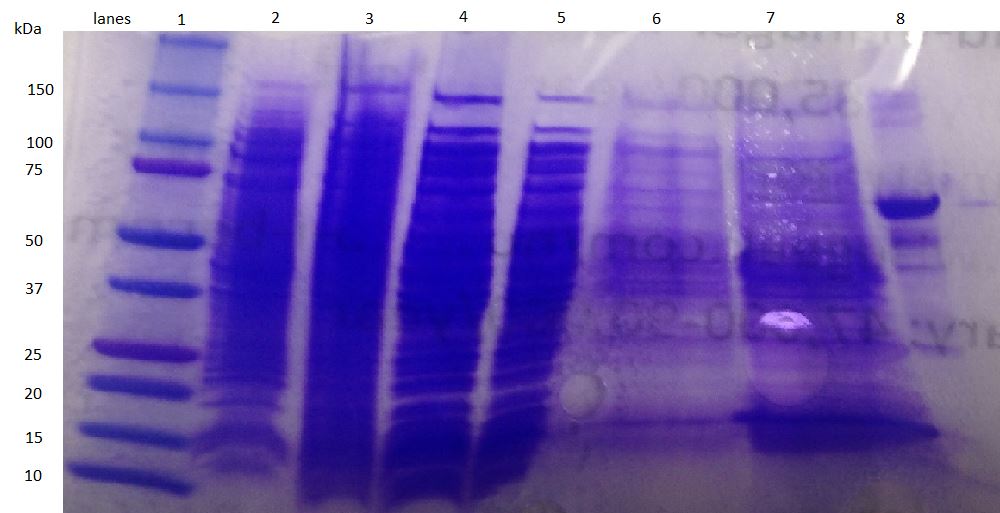
| + | *Here is our most recent attempt at expressing honeybee silk protein and the SDS PAGE results. We see a band at the expected size of 40 kDA! | ||
| + | [[File:UCLA honeybee Growth optimization 37C.jpg|none|thumb|300px|]] | ||
==<u>What we are working on now</u>== | ==<u>What we are working on now</u>== | ||
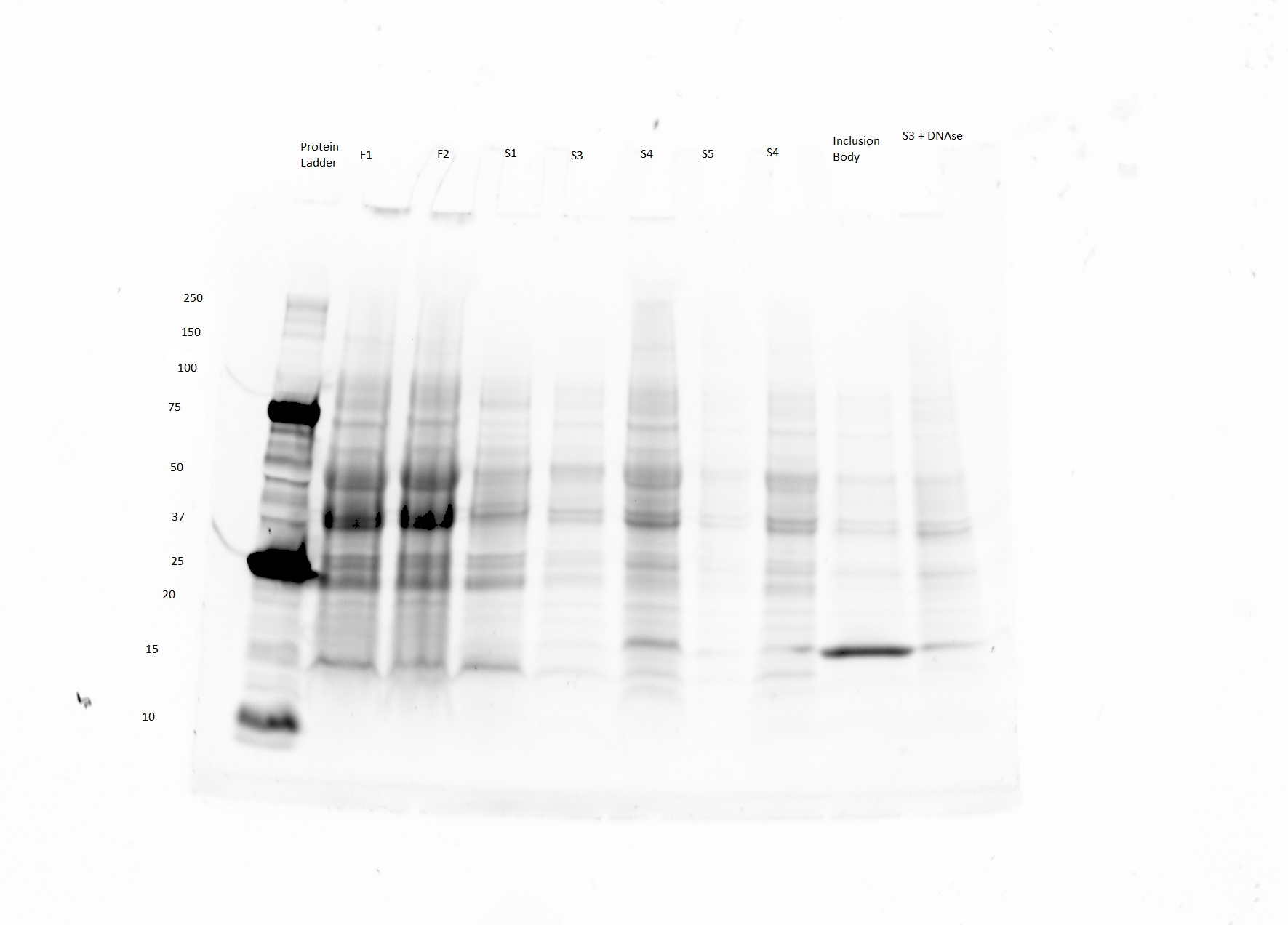
| − | We are currently | + | *We are currently optimizing our honeybee silk expression protocol. We are getting a band at the right size (40 kDa) on our SDS PAGE gels, but there is a fair amount of contamination. See example below (The purified product is in lane 7). |
| + | [[File:Honeybee BL21 honeybee SDS PAGE 8.7.15.JPG|none|thumb|300px|'''Fig. 1''' Expected size of product is 40 kDa (lane 6 and 7)). Lane 8 is BSA protein positive control, and lane 1 is Biorad dual color ladder. From lane 1-8 S1,F2,F3,S2,Final product 8/7, Final Product 7/29,BSA positive control, (see table)]] | ||
| + | *We are optimizing out cell lysis and purification protocol in the hopes of getting higher yield and a more pure protein product. | ||
| + | *In terms of producing fibers, we have "purified" honeybee product dissolved in SDS ready to go. We are waiting on the protein concentrators to arrive so that we can have a concentrated enough dope for spinning into fibers. | ||
| + | ** We hope that the protein concentration will also help to get rid of some of the contaminating proteins that are under the 30 kDa cutoff. | ||
==<u>Raw lab notebook entries</u>== | ==<u>Raw lab notebook entries</u>== | ||
| + | *April 28 - May 6 = cloning of [http://parts.igem.org/Part:BBa_K1763000 BBa_K1763000] and [http://parts.igem.org/Part:BBa_K1763001 BBa_K1763001 ] | ||
| + | *May 16-May 19 = Expressing Silk Protein in E. Coli. | ||
| + | *May 26-27 = Purifying Silk Protein and 1st SDS PAGE gel | ||
| + | *July 10-16 = Cloning Silk into Pet vector and preparing BL21 (DE3) competent cells | ||
| + | *July 17-23= Cloning Silk into BL21 (de3) cells. | ||
| + | *July 28-August 3rd = 1st expression in Bl21 (de3) and SDS PAGE results. | ||
| + | *August 7 = 2nd purfication and SDS of Honeybee silk in Bl21 cells along with SDS PAGE results. | ||
| + | *August 11 = BCA protein concentration assay on product from 8/7 | ||
| + | *August 12 = Optimization of growth conditions for Honeybee expression and SDS PAGE results. | ||
| + | *Sept 4,7 = SDS PAGE gels for different lysis protocols | ||
{{#calendar: year=2015 | month= apr | title = Team:UCLA/Notebook/Honeybee_Silk | query=preload=Template:UCLA}} | {{#calendar: year=2015 | month= apr | title = Team:UCLA/Notebook/Honeybee_Silk | query=preload=Template:UCLA}} | ||
{{#calendar: year=2015 | month= may | title = Team:UCLA/Notebook/Honeybee_Silk | query=preload=Template:UCLA}} | {{#calendar: year=2015 | month= may | title = Team:UCLA/Notebook/Honeybee_Silk | query=preload=Template:UCLA}} | ||
Latest revision as of 18:32, 18 September 2015
Contents
HONEYBEE SILK PROJECT
Goals
The goal of this project is to recombinantly express honeybee silk proteins, with the intention of creating synthetic honeybee silk fibers. We hope to develop protocols to efficiently produce the raw protein, process the protein into various materials, and characterize these materials. Furthermore, by conjugating the honeybee protein with other proteins, we aim to create honeybee silk materials with a wide array of properties and functionalities.
Achievements
As of 9/18 we are well on our way to establishing a strong foundation for this project. We have cloned five honey bee sequences into biobrick approved format, have expressed and purified honey bee silk protein. (For detailed achievements see below)
Cloning
We have successfully cloned and sequence verified five honeybee silk constructs into the igem psb1c3 plasmid backbone, in the correct biobrick format. A comprehensive list of our biobrick parts can be found here
- The first sequence is just the honeybee silk protein #3 in the psb1c3 backbone. The annotated sequence, along with further details on how we designed and cloned this biobrick can be found here. [http://parts.igem.org/Part:BBa_K1763000 BBa_K1763000]
- The second sequence is the same silk coding sequence, but with regulatory elements upstream for protein expression, including [http://parts.igem.org/Part:BBa_R0010 BBa_R0010 Promoter] and [http://parts.igem.org/Part:BBa_B0034 RBS BBa_B0034 ]. The sequence can be found here. [http://parts.igem.org/Part:BBa_K1763001 BBa_K1763001 ] along with further details on the design and cloning of this biobrick.
- The third sequence is a fusion protein between our honeybee silk and the Spycatcher, protein which has specific affinity for SpyCatcher. It also contains regulatory elements like a promoter and rbs for expression. You can view the sequence [http://parts.igem.org/Part:BBa_K1763008 BBa_K1763008 here .]
- The fourth sequence [http://parts.igem.org/Part:BBa_K1763007 BBa_K1763007 ] is our honeybee silk sequence with a T7 promoter upstream for effective protein expression in bacterial strains containing t7 polymerase.
- The fifth sequence, [http://parts.igem.org/Part:BBa_K1763015 BBa_K1763015] is our honeybee silk sequence+sfGFP with a T7 promoter and RBS upstream that can be used in bacterial strains that contain T7 polymerase.
Protein Expression
- We have just finished our first round of honey bee silk expression from [http://parts.igem.org/Part:BBa_K1763001 BBa_K1763001]
- Here is our first attempt at protein expression along with the gel.
- Unfortunately the purified protein that we see on the gel (should be in next to last lane on the right) doesn't seem to be the right size.
- We will try again using a T7 promoter and a slightly modified protein expression and purification protocol.
- Here is our most recent attempt at expressing honeybee silk protein and the SDS PAGE results. We see a band at the expected size of 40 kDA!
What we are working on now
- We are currently optimizing our honeybee silk expression protocol. We are getting a band at the right size (40 kDa) on our SDS PAGE gels, but there is a fair amount of contamination. See example below (The purified product is in lane 7).
- We are optimizing out cell lysis and purification protocol in the hopes of getting higher yield and a more pure protein product.
- In terms of producing fibers, we have "purified" honeybee product dissolved in SDS ready to go. We are waiting on the protein concentrators to arrive so that we can have a concentrated enough dope for spinning into fibers.
- We hope that the protein concentration will also help to get rid of some of the contaminating proteins that are under the 30 kDa cutoff.
Raw lab notebook entries
- April 28 - May 6 = cloning of [http://parts.igem.org/Part:BBa_K1763000 BBa_K1763000] and [http://parts.igem.org/Part:BBa_K1763001 BBa_K1763001 ]
- May 16-May 19 = Expressing Silk Protein in E. Coli.
- May 26-27 = Purifying Silk Protein and 1st SDS PAGE gel
- July 10-16 = Cloning Silk into Pet vector and preparing BL21 (DE3) competent cells
- July 17-23= Cloning Silk into BL21 (de3) cells.
- July 28-August 3rd = 1st expression in Bl21 (de3) and SDS PAGE results.
- August 7 = 2nd purfication and SDS of Honeybee silk in Bl21 cells along with SDS PAGE results.
- August 11 = BCA protein concentration assay on product from 8/7
- August 12 = Optimization of growth conditions for Honeybee expression and SDS PAGE results.
- Sept 4,7 = SDS PAGE gels for different lysis protocols
| April | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 27 | 28 | 29 | 30 | |||
| May | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | ||||
| 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| 18 | 19 | 20 | 21 | 22 | 23 | 24 |
| 25 | 26 | 27 | 28 | 29 | 30 | 31 |
| June | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 8 | 9 | 10 | 11 | 12 | 13 | 14 |
| 15 | 16 | 17 | 18 | 19 | 20 | 21 |
| 22 | 23 | 24 | 25 | 26 | 27 | 28 |
| 29 | 30 | |||||
| July | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 27 | 28 | 29 | 30 | 31 | ||
| August | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | |||||
| 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| 10 | 11 | 12 | 13 | 14 | 15 | 16 |
| 17 | 18 | 19 | 20 | 21 | 22 | 23 |
| 24 | 25 | 26 | 27 | 28 | 29 | 30 |
| 31 | ||||||
| September | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| 21 | 22 | 23 | 24 | 25 | 26 | 27 |
| 28 | 29 | 30 | ||||